Figure 5.
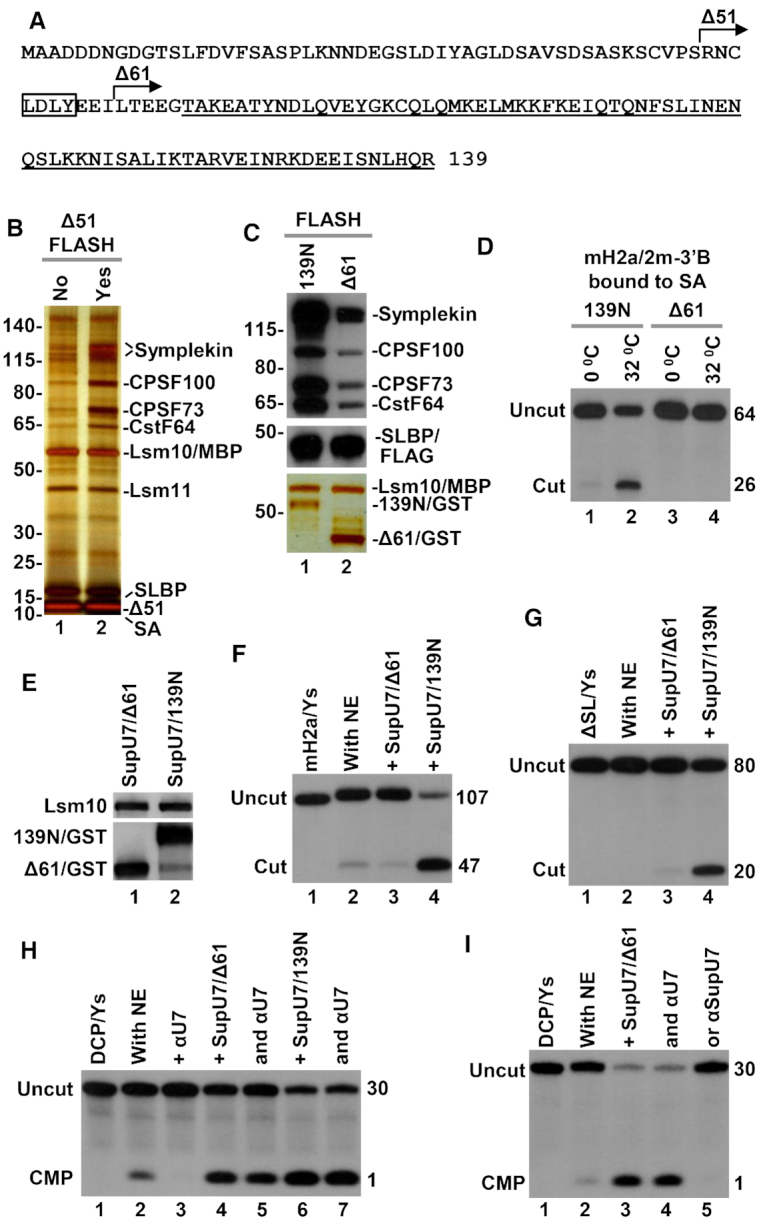
Role of FLASH in recruiting polyadenylation factors of the HCC to semi-recombinant holo U7 snRNP and its catalytic activity. (A) Amino acid sequence of the N-terminal region of FLASH. The essential LDLY motif is boxed and the start points of the Δ51 and Δ61 deletions are indicated with the arrows. The region that binds Lsm11 is underlined. (B) Proteins of mouse nuclear extract that associate with the in vitro assembled core U7 snRNP either lacking (lane 1) or containing (lane 2) recombinant Δ51 FLASH. The complexes were immobilized on SA beads via mH2a/2m-3′B pre-mRNA, as described in the legend for Figure 2A, and bound proteins were visualized by silver staining. (C) In vitro assembled core U7 snRNP was bound to either 139N FLASH or its inactive deletion mutant lacking the LDLY motif, Δ61. The resultant complexes were next bound in the presence of FLAG-tagged SLBP to radioactively labeled mH2a/2m-3′B pre-mRNA, immobilized on SA beads and incubated with mouse nuclear extract to reconstitute semi-recombinant holo U7 snRNP. The presence of the HCC components (symplekin, CPSF100, CPSF73 and CstF64), SLBP and each of the two forms of FLASH, each N-terminally tagged with GST, was monitored by Western blotting (top panel) and silver staining (bottom panel). (D) Small aliquots of immobilized processing complexes containing either 139N or Δ61 FLASH were incubated at 0°C and 32°C to evaluate their ability to support endonucleolytic cleavage of 5′-labeled mH2a/2m-3′B pre-mRNA. (E–I) In vitro assembled core SupU7 snRNP was bound to Δ61 or 139N FLASH. The complexes were purified by size exclusion chromatography, analyzed by Western blotting (panel E) and tested for the ability to support processing of H2a/Ys (panel F), ΔSL/Ys (panel G) and DCP/Ys (panels H and I) in a mouse nuclear extract.
