Figure 5.
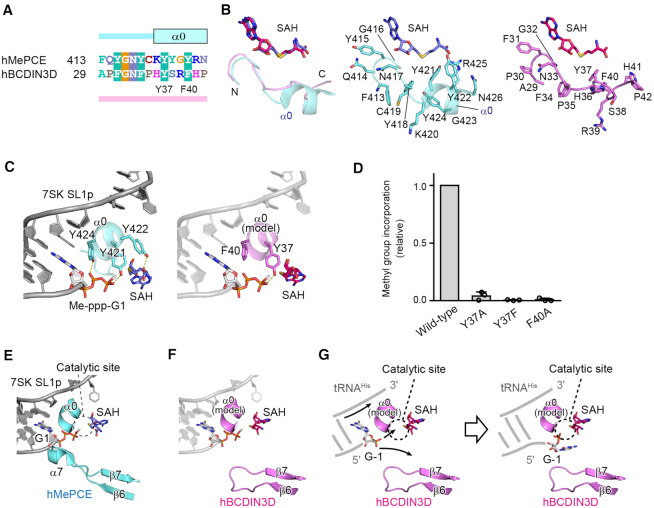
Recognition of the top part of the tRNAHis acceptor helix by BCDIN3D. (A) Sequence alignment of the N-terminal extended regions of the MTD of hMePCE and hBCDIN3D. In the structure of hMePCE complexed with SL1p, the N-terminal region forms an α-helix (α0). (B) Superimposition of the N-terminal region of hBCDIN3D (purple) complexed with SAH onto that of hMePCE (cyan) complexed with SL1p and SAH. The N-terminal region of BCDIN3D does not form an α-helix in the absence of RNA. Detailed views of the N-terminal regions of hMePCE (middle, cyan) and hBCDIN3D (right, purple). (C) Interactions between α0 and 7SK SL1p with the Me-ppp cap and SAH (left). The modeled α0 of BCDIN3D (purple) in the structure of hMePCE complexed with SL1p with the Me-ppp cap and SAH (right). Y37 in hBCDIN3D corresponds to Y421 and would interact with the phosphate group. (D) The activities of hBCDIN3D variants relative to the wild type (taken as 1.0) under the standard conditions. The reaction mixtures were incubated at 37°C for 1 h. Data are presented as mean ± SD of three independent assays. (E) Interactions of α7 between β6 and β7 of hMePCE and the 5′-end of 7SK SL1p with the Me-ppp cap and SAH. (F) The modeled α0 and the shorter loop between β6 and β7 of BCDIN3D with the SL1p, in the structure of hMePCE complexed with SL1p. The region between β6 and β7 of hBCDIN3D is shorter than the corresponding region of hMePCE, which forms α7 and interacts with the 5′-end of SL1p. (G) A possible mechanism of the α-phosphate methylation of tRNAHis by hBCDIN3D. Since an α-helix corresponding to α7 in hMePCE is absent in BCDIN3D, the 5′-monophosphate of G−1 of tRNAHis is deep within the catalytic pocket for 5′-phosphate methylation. α0 would wedge the G−1:A73 mispair at the top of the acceptor helix of tRNAHis.