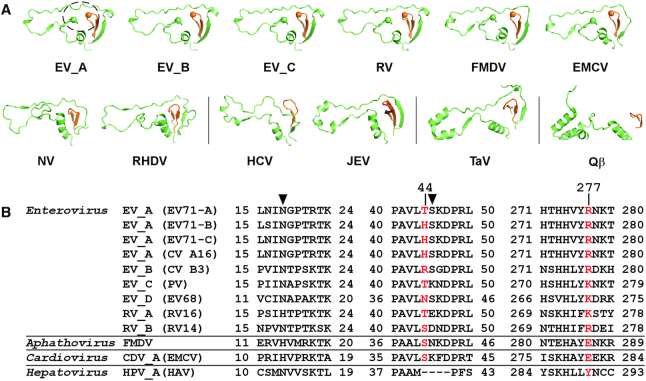
Figure 2.
Structure and sequence conservation analyses of the nucleobase-binding pocket. Virus abbreviations: FMDV–foot-and-mouth disease virus; EMCV–encephalomyocarditis virus; NV–norovirus; RHDV–rabbit hemorrhagic fever virus; HCV–hepatitis C virus; JEV–Japanese encephalitis virus; TaV–Thosea asigna virus; Qβ–bacteriophage Qβ; HAV–hepatitis A virus. (A) Side by side comparison of the index/middle (in green/orange) fingers subdomains of RdRPs from different positive-strand RNA viruses derived from a superposition (least square methods) of the highly conserved RdRP motif C residues. The dashed circle in the EV_A structure indicates the location of the nucleobase-binding pocket. PDB entries used: EV_A–6KWQ, EV_B–3N6L, EV_C–1RA6, RV–1TP7, FMDV–1U09 and EMCV–4NYZ for Picornaviridae (top row); NV–1SH0 and RHDV–1KHV for Caliciviridae; HCV–1C2P and JEV–4K6M for Flaviviridae; TaV–4XHI for Permutotetraviridae; Qβ–3MMP for Leviviridae. (B) Sequence alignment of three regions constituting the pocket (first two are part of the index finger and last one is part of the middle finger) for RdRPs from the Enterovirus genus and three representative RdRPs from other genera of the Picornaviridae. Residues equivalent to the nucleobase stacking H44 and R277 in EV_A RdRP are shown in red, while those equivalent to the hydrogen bonding N18 and S45 are indicated by solid triangles.