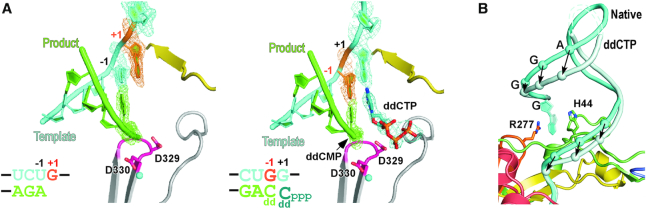
Figure 3.
A comparison of the native and ddCTP-derived EC structures. (A) The EC structures are shown in cartoon representations with composite SA omit electron density map (contoured at 1.2 σ) overlaid for the native (left) and ddCTP-derived ECs. For clarity, the majority of the polymerase and RNA up- and downstream of the active site are not shown. Except for the templating +1 nt in the native complex and the −1 nt in the ddCTP complex are shown in orange to correlate the nucleic acids translocation, the coloring scheme is as in Figure 1. Magnesium ions are shown as cyan spheres. (B) A comparison of the downstream RNA of the two EC structures. The arrows indicate the movement of the backbone phosphates of equivalent nucleotides from the native (cyan) conformation to the ddCTP (light cyan) conformation. Dashed lines indicate the likely path of the disordered RNA in the ddCTP complex. RdRP structure is only shown for the native complex for clarity.