Figure 2.
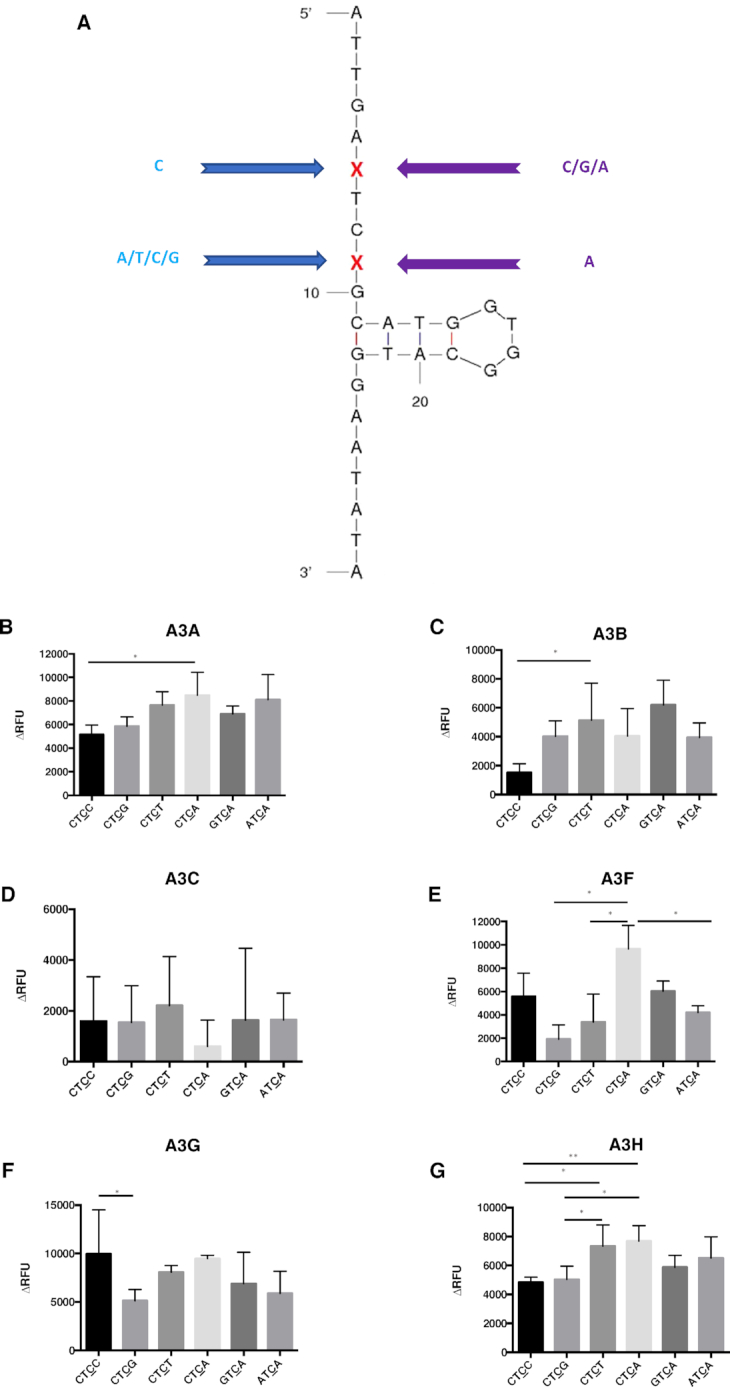
APOBEC3 proteins have different preferences with oligonucleotides having different flanking bases on either side of the target site. (A) Oligonucleotide sequence and predicted secondary structure. Cytidine was chosen for the 5′ flanking base of ‘TC’ motif, while the 3′ flanking base is either adenine, cytidine, guanine, or thymidine. In contrast, adenine was chosen for the fixed 3′ flanking base of ‘TC’ motif, while the 5′ flanking base is either adenine, cytidine or guanine. The same sequence (backbone) as the ‘open’ oligonucleotides was used for all the substrates besides the modifications of the flanking bases. (B–G) APOBEC3 protein activity with different oligonucleotide substrates. Comparison of APOBEC3 activity was determined among the oligonucleotides CTCC, CTCG, CTCT, CTCA, GTCA and the ATCA open oligonucleotide. Results are shown as the average ± standard deviation of 3 independent experiments. The asterisks and brackets above any two bars indicate the significant difference between experiments. Significance was analyzed by using a one-way ANOVA with significance defined as P ≤ 0.05. Experiments that were not significant or significant are indicated as follows: ‘ns’ = not significant; ‘**’ = P ≤ 0.01; ‘*’ = P ≤ 0.05.
