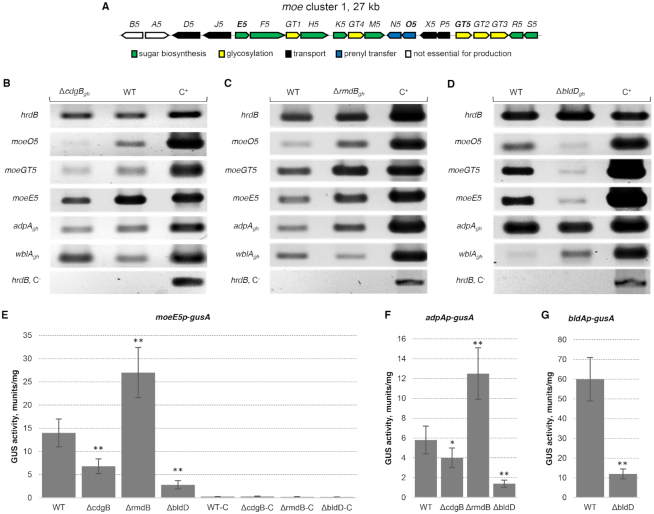
Figure 2.
Deletion of cdgBgh,rmdBgh and bldDgh strongly influences expression of genes essential for moenomycin production. (A) Genetic organization of moe cluster 1. (B) Comparison of ΔcdgBgh and ATCC14672 transcriptional profiles. (C) Comparison of ΔrmdBgh and ATCC14672 transcriptional profiles. (D) Comparison of ΔbldDgh and ATCC14672 transcriptional profiles. The expression of tested genes was analyzed in 48 h cultures grown in TSB; 200 ng of RNA sample were used per reaction; C+, positive control (genomic DNA of ATCC14672 strain). Attempts to synthesize hrdB from RNA without pretreatment with RT served as negative controls (marked as C−). Total RNA samples were isolated from three independent biological replicates. The images represent the typical result of three independent RT-PCR experiments. (E) Transcriptional activity of the moeE5 promoter in Streptomyces ghanaensis strains. (F) Transcriptional activity of the adpAgh promoter in S. ghanaensis strains. (G) Transcriptional activity of the bldAgh promoter in S. ghanaensis strains. WT-C, ΔcdgB-C, ΔrmdB-C and ΔbldD-C: control strains carrying promoterless vector pGUS. Cultures and subsequent β-glucuronidase assays were performed in triplicate. Values were normalized to equal amounts of dry biomass. Error bars, ±2 SD. Significance of difference in the transcriptional activities of tested promoters was calculated by a two-tailed t-test. Asterisks indicate the significance value (∗P < 0.05, ∗∗P < 0.01).