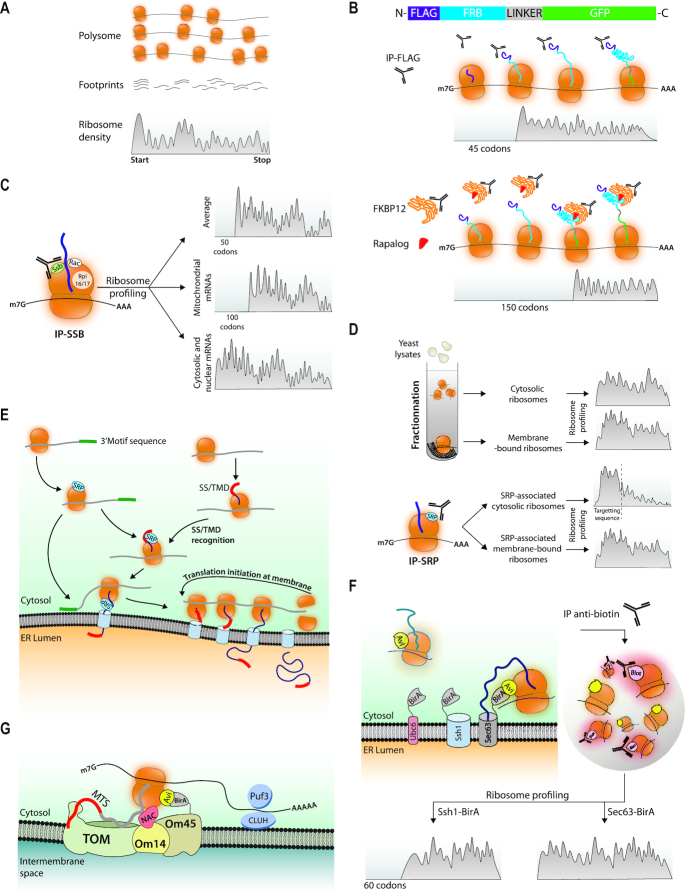
Figure 3.
Ribosome profiling associated with selective purification steps determines timing of co-translational folding, chaperone binding and targeting. (A) Ribosome profiling. This is a technique in which total polysomes prepared from a given sample are treated with RNAse to generate monosomes from which the ribosome-protected fragments called footprints are purified and used to make a library that is sequenced. Each fragment is defined by the position of the ribosome P site and the total amount of footprints aligning to each codon normalized to the total complexity of the library (RPKM, y axis) are aligned to reference sequences (x axis). The hypothetical footprints on one ORF are depicted. (B) Ribosome profiling to evaluate co-translational protein folding from cells expressing Flag-FRB-GFP. Upper panel: Ribosome footprints recovered in a Flag immunoprecipitate map to the ORF starting at a position corresponding to ribosomes that had just synthesized enough of the nascent chain to expose the Flag epitope outside of the ribosome tunnel. Lower panel: Ribosome footprints recovered in a FKBP12 immunoprecipitate in presence of a Rapalog map to the ORF starting at a position corresponding to ribosomes that had just synthesized enough of the nascent chain to expose FRB outside of the ribosome tunnel. (C) Ribosome profiling to evaluate co-translational chaperone recruitment. Ssb was immunoprecipitated and the ribosome footprints recovered in the immunoprecipitate were mapped. A large majority, but not all, mRNAs were recovered and the patterns of the ribosome footprints depended upon the category of the proteins encoded by these mRNAs. (D) Ribosome profiling to evaluate Srb targeting to the ER. Upper panel: Cytosol or membranes were fractionated from total extracts and the ribosome footprints recovered in the 2 fractions were mapped. Footprints tended to be distributed throughout the ORFs. Then Srb was immunoprecipitated and the ribosome footprints recovered in the immunoprecipitate were mapped. Lower panel: The Srb-engaged ribosomes from the cytosolic pool showed a gradual depletion of footprints at a position corresponding to exposure of the signal sequence outside of the ribosome tunnel. The Srb-engaged ribosomes from the membrane pool showed mostly distribution throughout the ORF. (E) Proximity labeling at the ER. The Sec61 or Ssh1 transolocon subunits or the C-terminal tail-anchor of Ubc6 were fused to BirA in cells where a ribosomal subunit of either the 40S or of the 60S subunit was fused to an Avi-tag. Ribosomes in proximity to the BirA tagged proteins get biotinylated and can be purified to then map ribosome footprints. (F) Updated model of protein targeting to the ER. The mRNAs encoding proteins with or without a signal sequence recruit SRP before any signal sequence is exposed out of the ribosome tunnel, and 3′UTR sequences can contribute to SRP recruitment as well targeting to the ER (82). SRP is then poised to associate with the signal sequence and dock the RNCs to its receptor and transfer them to the translocon. mRNAs targeted to the ER stay for new rounds of translation. (G) Proximity labeling to investigate targeting of ribosomes to the mitochondrion. BirA was fused to Om45. Om45 interacts with Om14 that acts as a receptor for ribosomes translating mitochondrial proteins via the NAC chaperone, leading to nascent chain transfer to the TOM complex. Targeting to the mitochondria has been attributed on one hand to the nascent chain MTS (mitochondrial targeting sequence) and on the other to the mRNA via RNA binding proteins such as Puf3 in yeast or CLUH in mammalian cells (83–87)).