Figure 1.
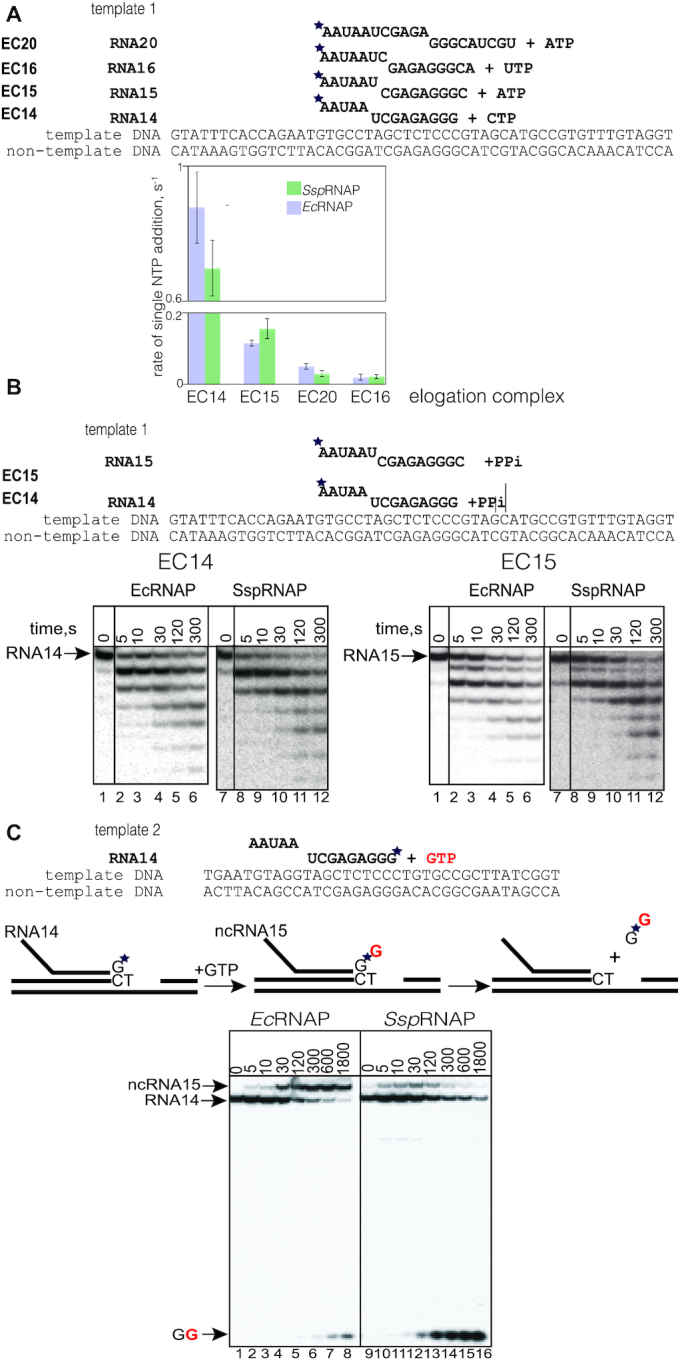
Rates of correct and incorrect substrate incorporation, and rates of pyrophosphorolysis are similar between Ssp and EcRNAPs. (A) Rates of incorporation of correct NTPs are similar for SspRNAP and EcRNAP. Rates of incorporation (s−1) of the single correct (1 μM) substrates by SspRNAP and EcRNAP were compared in elongation complexes with 14 nt, 15 nt, 16 nt and 20 nt long RNA (EC14, EC15, EC16 and EC20) on template 1, are presented as bars on the graph. Scheme and sequence of RNA and DNA strands are above the graph. The elongation complexes were obtained by extension of initial RNA13, labelled with 32P at the 5′-end as indicated by the asterisk, upon NTPs addition. Error bars represent standard deviations from triplicate experiments. (B) Rates of pyrophosphorolysis are similar for SspRNAP and EcRNAP. Kinetics of pyrophosphorolysis in EC14 and EC15 on template 1 using 250 μM pyrophosphate. (C) SspRNAP misincorporates substrates with same efficiency as EcRNAP, and proofreads a mistake faster. Kinetics of misincorporation by EcRNAP and SspRNAP, representative gel for misincorporation reaction of GTP instead of template-dictated ATP in EC14 on template 2. Schematics above the gel show elongation complex and reactions of misincorporation and subsequent hydrolysis with dinucleotide product release. Asterisk indicates that RNA is labelled at the 3′ end, which allows monitoring of misincorporation and proofreading simultaneously. Initial 14nt RNA (RNA14) after GTP misincorporation elongates to ncRNA15 (non-correct), then the 3′ incorrect dinucleotide piece of the transcript (GG) is cleaved out.
