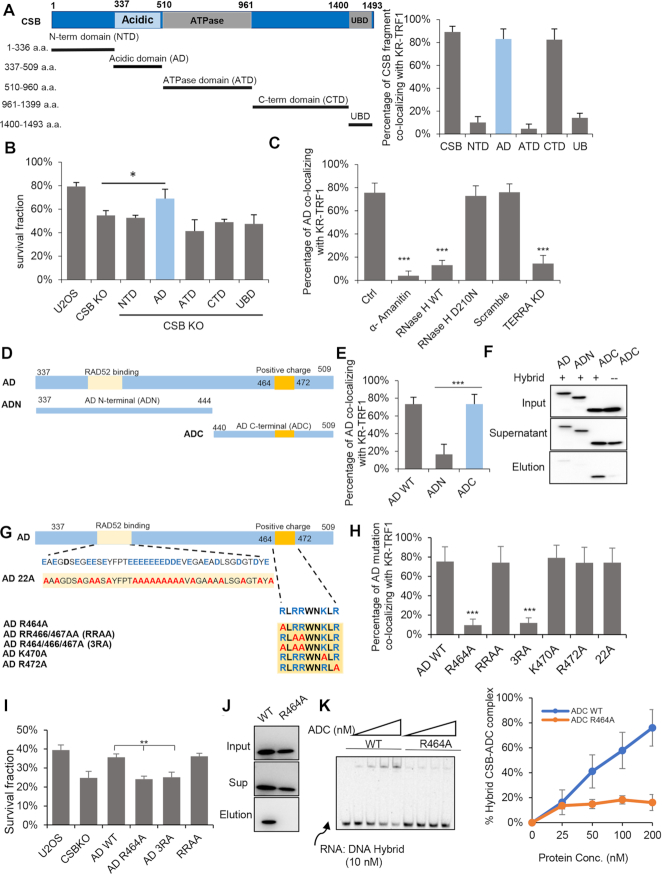
Figure 3.
R464 of CSB contributes to DNA: RNA hybrid binding. (A) Schematic diagram of CSB fragments. The left panel is the quantification of co-location frequency of CSB fragments to KR-TRF1in U2OS cells. (B) Cell survival of CSB WT and KO cells expressing indicated fragments of CSB with 1 h light activation of KR-TRF1 in U2OS cells. n = 3, unpaired t-test, error bars represent SD, *P < 0.05 (C) GFP-AD foci co-localization with KR-TRF1 in U2OS cells treated with α-amanitin (100 μg/ml, 2 h), overexpressed with HA-RNaseH WT, D210N mutant or TERRA depleted with LNA. (D) Schematic diagram of CSB-ADN and ADC. (E) AD WT, ADN and ADC co-localization with KR-TRF1 after light activation. (F) The affinity of GFP-AD, GFP-ADN, and GFP-ADC to DNA: RNA hybrid by biotin-labeled hybrid pulldown using cell lysate from U2OS CSB KO cells. (G) Schematic diagram of CSB-AD 22A, R464A, RRAA, 3RA, K470A and R472A. (H) The recruitment of CSB-AD mutants to KR-TRF1 after light activation. (I) Colony formation assays for U2OS CSB KO cells expressing GFP-AD WT, AD R464A, AD3RA, and AD RRAA after inducing ROS damage by KR-TRF1 with 1.5 light activation. n = 3, unpaired t-test, error bars represent SD, **P < 0.01 (J) The affinity of GFP-ADC and GFP-ADC R464A to DNA: RNA hybrid by biotin-labeled hybrid pulldown using cell lysate from U2OS CSB KO cells. (K) DNA:RNA hybrids EMSA assay of purified CSB-ADC WT and R464A protein. For (C), (E) and (H), Mean values with SD from 50 cells in three independent experiments are given. P-value is calculated by unpaired t-test. ***P < 0.001.