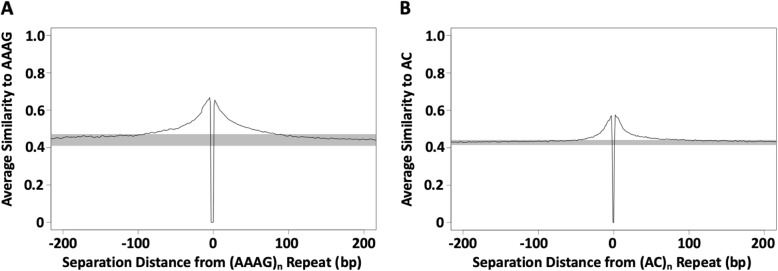
Fig. 3.
Decay of sequence similarity with distance from perfect SSR repeats. Average similarities were calculated for short segments within 200 bp of perfect SSR repeats with a given motif. Similarity was measured as the proportion of identical nucleotides at each position for a segment of the same length and read direction as the repeated motif shown, (AAAG)n in a, (AC)n in b. For example, a segment reading “ATAG” would have a similarity of 0.75 with the repeat motif “AAAG”. Average similarities were calculated for segments beginning at every nucleotide separation distance within 200 bp of the perfect repeat beginning or end. The black line shows the average similarity to each repeat, while the gray box shows a range of 3 standard deviations from the mean similarities calculated in 700 bp windows from 300 to 1000 bp away from both ends of the perfect repeat loci. The dips near x = 0 reflect that a non-motif base must precede and follow the perfect region of the repeat at the start and end of the perfectly repeated segment