Figure 2.
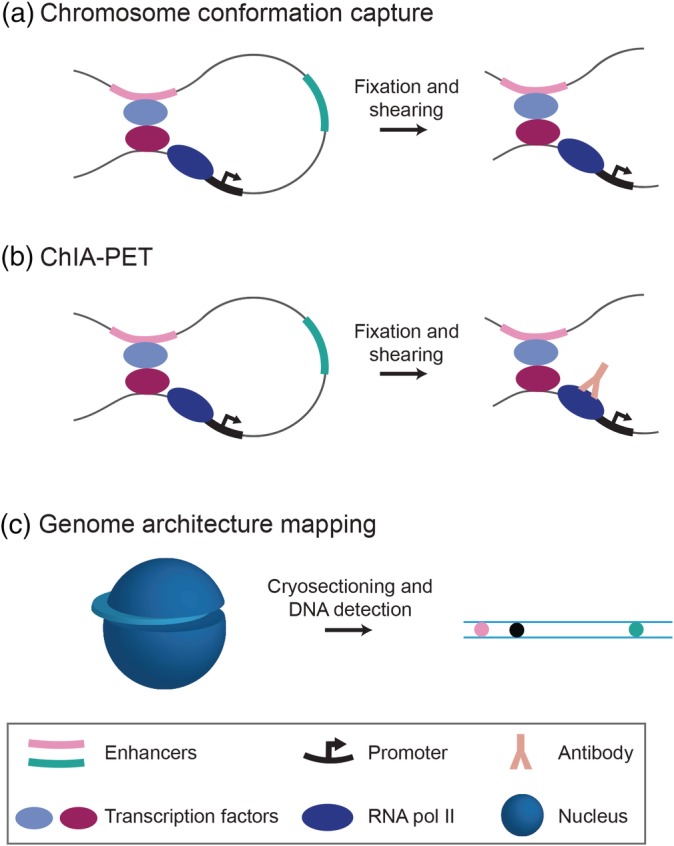
Genome‐wide methods to identify putative enhancer–promoter interactions. Distal enhancers (shown in pink) may be brought within close proximity to the promoters they regulate (shown in black) through the formation of chromatin loops. Looping is facilitated by protein–protein interactions of transcription factors that assemble at promoters and enhancers. The basic principles of methods to detect chromatin interactions are shown. Both (a) chromatin conformation capture (3C)‐based methods, including Hi‐C, and (b) chromatin interaction analysis with paired‐end tag sequencing (ChIA‐PET) preserve and detect chromatin interactions through crosslinking, fragmentation, and proximity ligation followed by high‐throughput sequencing. ChIA‐PET includes a chromatin immunoprecipitation step, often using antibodies targeting RNA pol II (shown), to enrich for complexes containing promoters. (c) Genome architecture mapping captures the distance between genomic loci by cryosectioning and laser microdissection (which allow spatial information to be preserved) followed by DNA sequencing
