Figure 3.
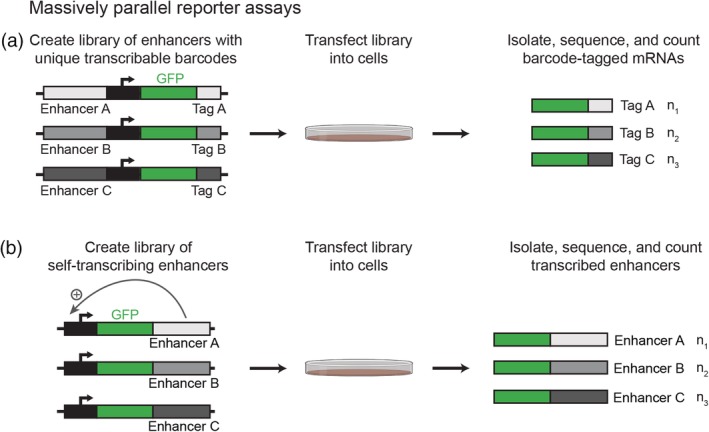
In vitro massively parallel reporter assays (MPRAs) to study enhancer function. MPRAs utilize advances in oligonucleotide synthesis and sequencing technology to test large libraries of candidate enhancer sequences for function. (a) MPRA libraries typically contain candidate enhancer sequences upstream of a minimal promoter and reporter gene. A transcribable barcode unique to each candidate enhancer is placed in the 3′ untranslated region of the reporter gene, allowing active enhancers to be identified by sequencing barcode‐tagged mRNAs. (b) Self‐transcribing active regulatory region sequencing (STARR‐seq) is a variation of the MPRA that exploits the characteristic that enhancers can function independently of their relative positions. In STARR‐seq, candidate enhancer sequences are placed downstream of a minimal promoter and reporter gene, allowing active enhancers to transcribe themselves. Figure created with BioRender
