Figure 4.
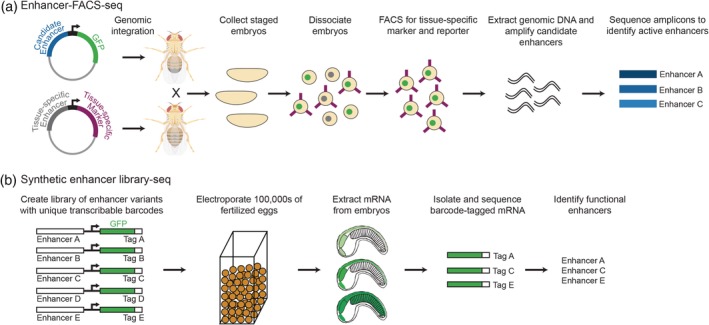
In vivo massively parallel reporter assays to study enhancer function. (a) Enhancer‐FACS‐seq identifies active, tissue‐specific enhancers in whole Drosophila embryos. Two flies are crossed, the first carrying a candidate enhancer sequence driving GFP expression, and the second expressing a cell surface marker in a tissue‐specific manner. Embryos resulting from this cross are dissociated and FACS sorted for the tissue‐specific cell surface marker and GFP. Genomic DNA can then be extracted from sorted cells to identify enhancers that are active in the specific tissue. (b) Synthetic enhancer library‐sequencing (SEL‐seq) allows millions of enhancer variants to be tested for function in whole Ciona embryos. Synthetic enhancer variants are attached to a minimal promoter, GFP coding sequence, and a unique transcribable barcode. The enhancer library is electroporated into hundreds of thousands of fertilized Ciona eggs, which are then allowed to develop until the desired developmental stage. Barcode‐tagged mRNA can then be isolated and sequenced to identify active enhancer variants, providing insight into which sequences within an enhancer are important for function. Figure created with BioRender
