Figure 5.
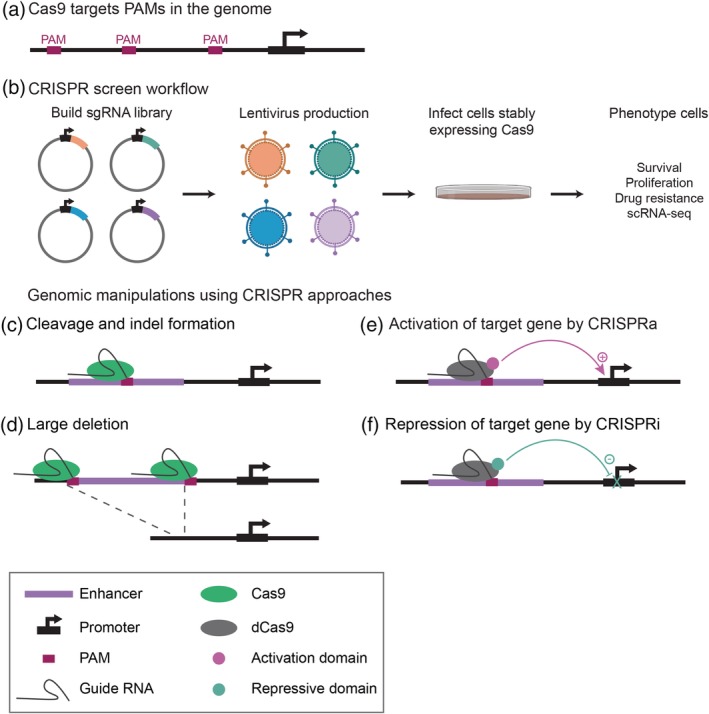
CRISPR‐based methods to identify and study candidate enhancers. (a) Any genomic region containing a protospacer‐adjacent motif (PAM) can be targeted by CRISPR/Cas9. (b) The workflow for high‐throughput CRISPR screens includes building a library of sgRNAs, cloning the library into lentiviral vectors, infecting cells that stably express Cas9 or one of its variant forms, and phenotyping cells. The sgRNA(s) carried by cells of a given phenotype can easily be identified by sequencing, and can give insight into candidate cis‐regulatory regions that affect phenotype. CRISPR screens can utilize (c) one sgRNA to mutate a particular locus, or (d) two sgRNAs to delete a genomic region. In both cases, DNA cut by Cas9 will be repaired by the error‐prone non‐homologous end joining (NHEJ) pathway, creating indels. Variant forms of Cas9 that are used in high‐throughput screens include (e) CRISPRa to activate transcription and (f) CRISPRi to repress transcription. Both CRISPRa and CRISPRi utilize catalytically dead Cas9 fused to an effector protein and impact gene expression through chromatin remodeling. Figure created with BioRender
