Figure 2.
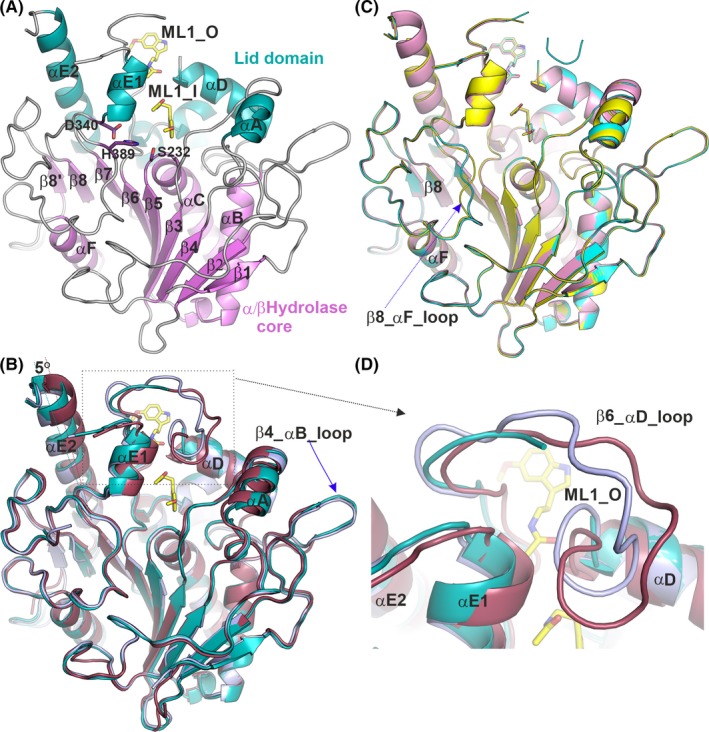
Cartoon representations of Notum structures. A, Notum‐melatonin complex structure. The conserved α/β‐hydrolase core structures are coloured in violet, the lid domain in teal and loops in grey. Melatonin is shown as yellow sticks. The enzyme catalytic triad side chains are shown as violet sticks. B, Alignment of the Notum‐melatonin complex (PDB code 6TR5), in teal colour) with two apo structures; one in the open conformation (PDB code 4UZ1, in light blue) and one in the closed conformation (dark red; PDB code 4UYU). C, Alignment of structures of Notum bound to melatonin (yellow; PDB code: 6TR5), N‐[2‐(5‐fluoro‐1H‐indol‐3‐yl)ethyl]acetamide (cyan; PDB code 6TR7) and N‐acetylserotonin (magenta; PDB code: 6TR7). D, Close up of the β6_αD loop region from B showing movement or disorder in apo and melatonin‐bound structures
