Figure 2.
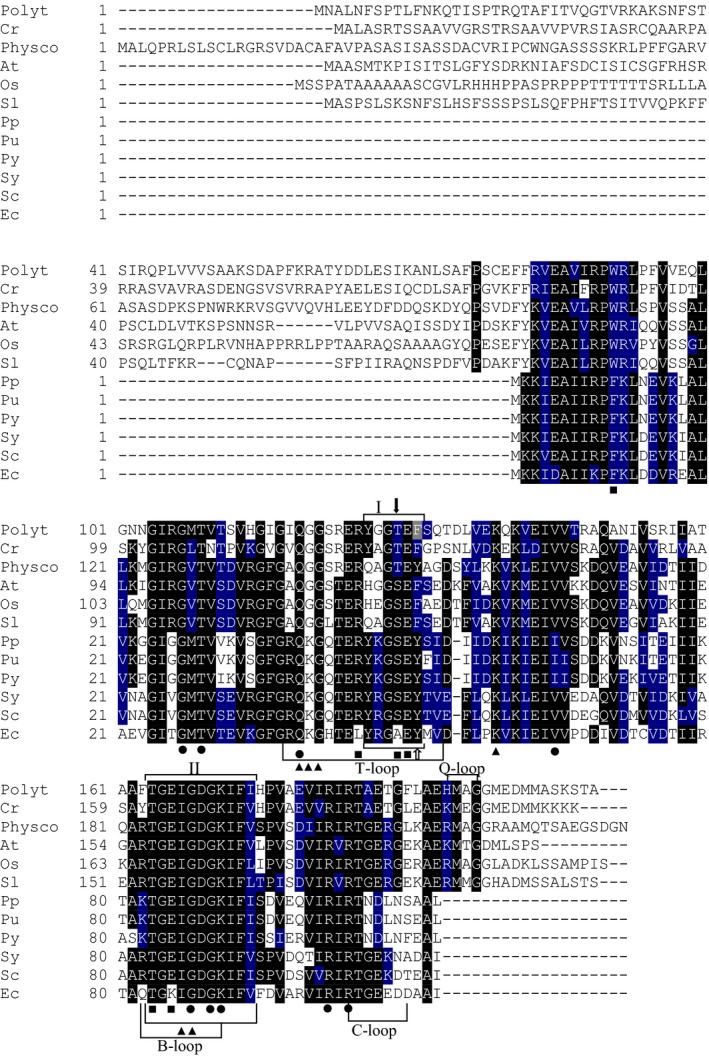
Multiple amino acid sequence alignment of PII proteins. The protein sequences were derived from NCBI database. The sequences are derived from PII polypeptides of the nonphotosynthetic alga Polytomella parva (Polyt), green photosynthetic alga Chlamydomonas reinhardtii (Cr; XP_001703658.1), land plants Physcomitrella patens (Physco; BAF36548.1), Arabidopsis thaliana (At; NP_192099.1), Oryza sativa Japonica (Os; Os05g0133100) and Solanum lycopersicum (Sl; AAR14689.1), red algae Porphyra purpurea (Pp; NP_053864.1), Porphyra umbilicalis (Pu; AFC39923.1) and Pyropia yezoensis (Py; AGH27579.1), cyanobacteria Synechococcus elongatus PCC 7942 (Sy; P0A3F4.1), Synechocystis sp. PCC 6803 (Sc; CAA66127.1) and Escherichia coli (Ec; CAQ32926.1). All the indicated regions and residues have been characterized in previous work 17, 24, 25, 26, 27. The regions referring to T‐, B‐, C‐ and Q‐loops are indicated 17. Highlighted residues in black are invariant in at least 55% of aligned PIIs proteins. Amino acids in blue represent similar residues. Boxs I and II indicate PII signature patterns. The positions of known PIIs post‐translational modification sites: the phosphorylation site in cyanobacterial S. elongatus PII (S49) and the uridylation site in E. coli PII (Y51) are indicated by solid black and white arrows, respectively. The amino acid residues involved in binding of ATP (●), NAGK (■) and 2‐OG (▲) are indicated 24, 25, 26, 27. The alignment was done using the ClustalW program and manually refined.
