Figure 3.
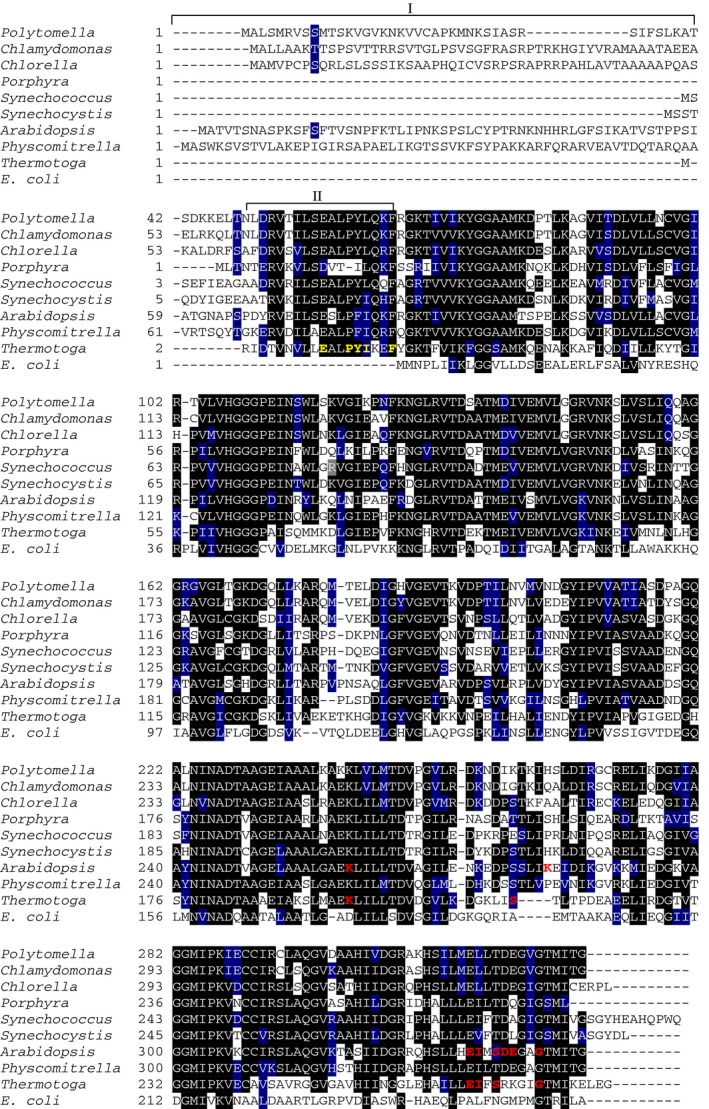
Multiple amino acid sequence alignment of NAGK proteins. The NAGK protein sequences were derived from UniProt database. The sequences are derived from NAGK polypeptides from nonphotosynthetic alga Polytomella parva (A6XGV3), green photosynthetic alga Chlamydomonas reinhardtii (A8HPI1) and Chlorella variabilis (E1ZQ49), land plants Physcomitrella patens (A0JC02) and Arabidopsis thaliana (Q9SCL7), red algae Porphyra purpurea (P69365), cyanobacteria Synechococcus elongatus PCC 7942 (Q6V1L5) and Synechocystis sp. PCC 6803 (P73326), and bacteria Thermotoga maritima (Q9X2A4) and Escherichia coli (P0A6C8). Highlighted residues in black are invariant in at least 55% of aligned NAGK proteins. Amino acids in blue represent similar residues. Box I refers to plastid‐targeting signal peptides sequence (ChloP server). Box II indicates an N‐terminal signature extension of Arg‐sensitive NAGK proteins, which is absent in Arg‐insensitive E. coli NAGK 28. In Box II, the previously identified signature sequence of Arg‐sensitive NAGK from Thermotoga maritima is highlighted in yellow, which is involved in forming the allosteric Arg binding site 28. Amino acid residues directly involved in allosteric Arg binding are highlighted in red and are deduced from known structures of NAGK: Arg complexes from Thermotoga maritima NAGK (PDB: http://www.rcsb.org/pdb/search/structidSearch.do?structureId=2BTY) 28 and Arabidopsis thaliana (PDB: http://www.rcsb.org/pdb/search/structidSearch.do?structureId=2RD5) 26. The alignment was done using the ClustalW program and manually refined.
