Figure 3.
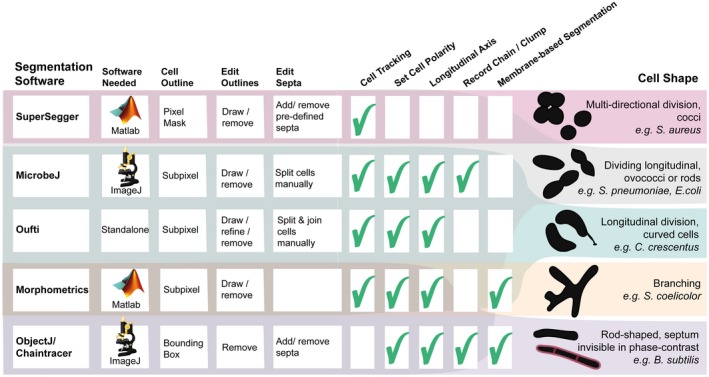
Overview of the functionality of five programs compatible with BactMAP. Of the five tested programs, three are MATLAB‐Based (SuperSegger, Morphometrics and Oufti) and two are ImageJ Plugins (MicrobeJ, ObjectJ). While Oufti is MATLAB‐Based, it comes as a standalone program for 64x operating systems. In addition to measuring the outlines, Oufti, MicrobeJ, Morphometrics and SuperSegger can track cells over time and provide information on growth speed and cell genealogy. Oufti, MicrobeJ, Morphometrics and ObjectJ estimate the cell length and curvature over the longitudinal axis. MicrobeJ offers a range of options for detection and counting of cell chains and clumps, while both MicrobeJ and ObjectJ offer options to detect cell features such as curvatures or invaginations as specified by the user. Finally, both SuperSegger and MicrobeJ give users the option to group cells based on user‐specified cell features. All programs offer some options for manual editing of the results. In Oufti, a user can split or join cells, delete cells and draw new cell outlines. In Morphometrics, MicrobeJ and ObjectJ, it is also possible to delete or add cells. For both Morphometrics and Oufti, it is not possible to move septa to a manually chosen subcellular location. In MicrobeJ this is possible, just as ObjectJ's ChainTracer allows users to check, add and delete detected septa manually. Also in SuperSegger, it is possible to delete cells, but it is only possible to delete or add septa on pre‐calculated positions. The right panel shows which program performs cell segmentation best on which kind of shaped cells in our experience [Colour figure can be viewed at https://www.wileyonlinelibrary.com]
