Abstract
DNA double-strand breaks are genotoxic lesions whose repair can be templated off an intact DNA duplex through the conserved homologous recombination (HR) pathway. Because it mainly consists of a succession of non-covalent associations of molecules, HR is intrinsically reversible. Reversibility serves as an integral property of HR, exploited and tuned at various stages throughout the pathway with anti- and pro-recombinogenic consequences. Here, we focus on the reversibility of displacement loops (D-loops), a central DNA joint molecule intermediate whose dynamics and regulation have recently been physically probed in somatic S. cerevisiae cells. From homology search to repair completion, we discuss putative roles of D-loop reversibility in repair fidelity and outcome.
Keywords: D-loop, Homologous recombination, Helicase, Homology search, Genomic stability, Crossover
Homologous recombination is a succession of metastable intermediates
Homologous recombination (HR) is a conserved pathway for the repair of DNA double-strand breaks (DSBs) (Fig. 1). Nucleolytic resection of the DSB generates long 3′-protruding single-stranded DNA, uncovering several hundreds to thousands of nucleotide sequence around the break site (Symington 2016). This sequence information is harnessed by a multivalent helical filament of RecA (in bacteria), RadA (in archea) or Rad51 (in eukaryotes, with additional cofactors) using base-pairing principles for the dynamic genome-wide search for a homologous match (Bell and Kowalczykowski 2016; Bordelet and Dubrana 2019; Zimmer and Fabre 2019). Once significant homology is identified by the nucleoprotein filament (NPF), DNA strand invasion can occur, which results in a DNA joint molecule called a displacement loop (D-loop) (Wright et al. 2018). A D-loop contains a B-form heteroduplex DNA (hDNA) region formed between the broken molecule and a complementary strand in another duplex DNA, a displaced strand, and junctions at both ends (Fig. 1). If incorporated in the hDNA, the 3′ end of the broken molecule can be extended by a DNA polymerase using the complementary strand as a template to generate an extended D-loop. This step restores the disrupted sequence information and provides opportunities for annealing or invasion to the other side of the DSB for repair completion by second end capture. The various HR sub-pathways and their associated repair outcomes branch-out depending on the processing of extended D-loops [for more information see (Mehta and Haber 2014; Piazza and Heyer 2019)]. In somatic cells, the main pathway is termed synthesis-dependent strand annealing (SDSA) (Mehta and Haber 2014). SDSA entails the disruption of the extended D-loop and engagement of the second end of the DSB (Fig. 1).
Fig. 1.
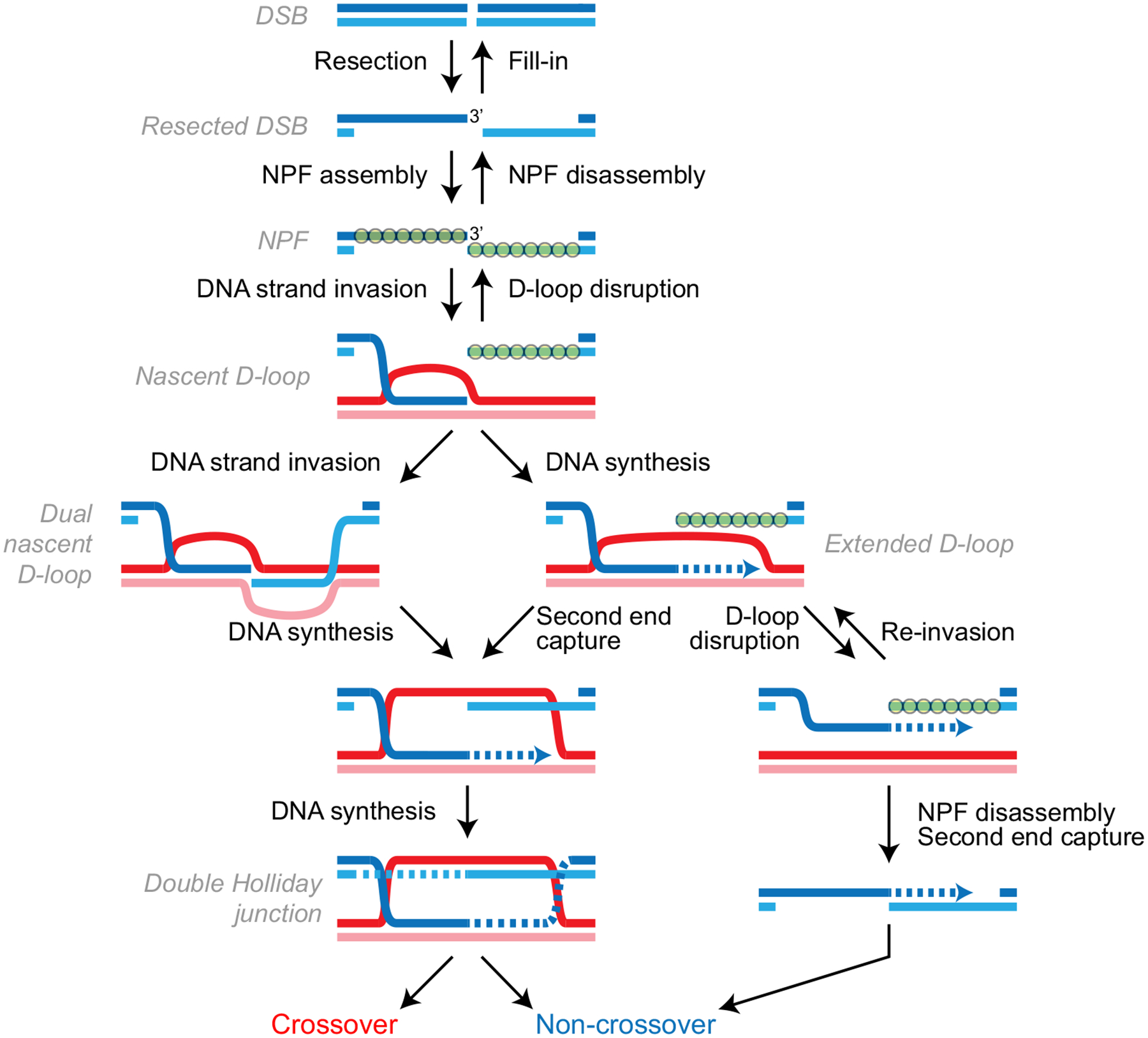
Homologous recombination is a complex pathway involving multiple meta-stable reversible intermediates. Resection can be reverted by fill-in by Polα-dependent DNA synthesis (Mirman et al. 2018), the NPF can be disassembled by the action of helicases and ubiquitin ligases (Branzei and Szakal 2017), and D-loops can be disrupted (see text). Second end capture can be accomplished by two different mechanisms, annealing of two single-strand DNAs or invasion of duplex DNA of the second end (past the resection junction) by the extended first end (not shown)
D-loops are decision nodes of HR, acted upon by multiple protein players
D-loops are pivotal intermediates of the HR pathway that are substrate for three main classes of enzymes (Fig. 2a):
Fig. 2.
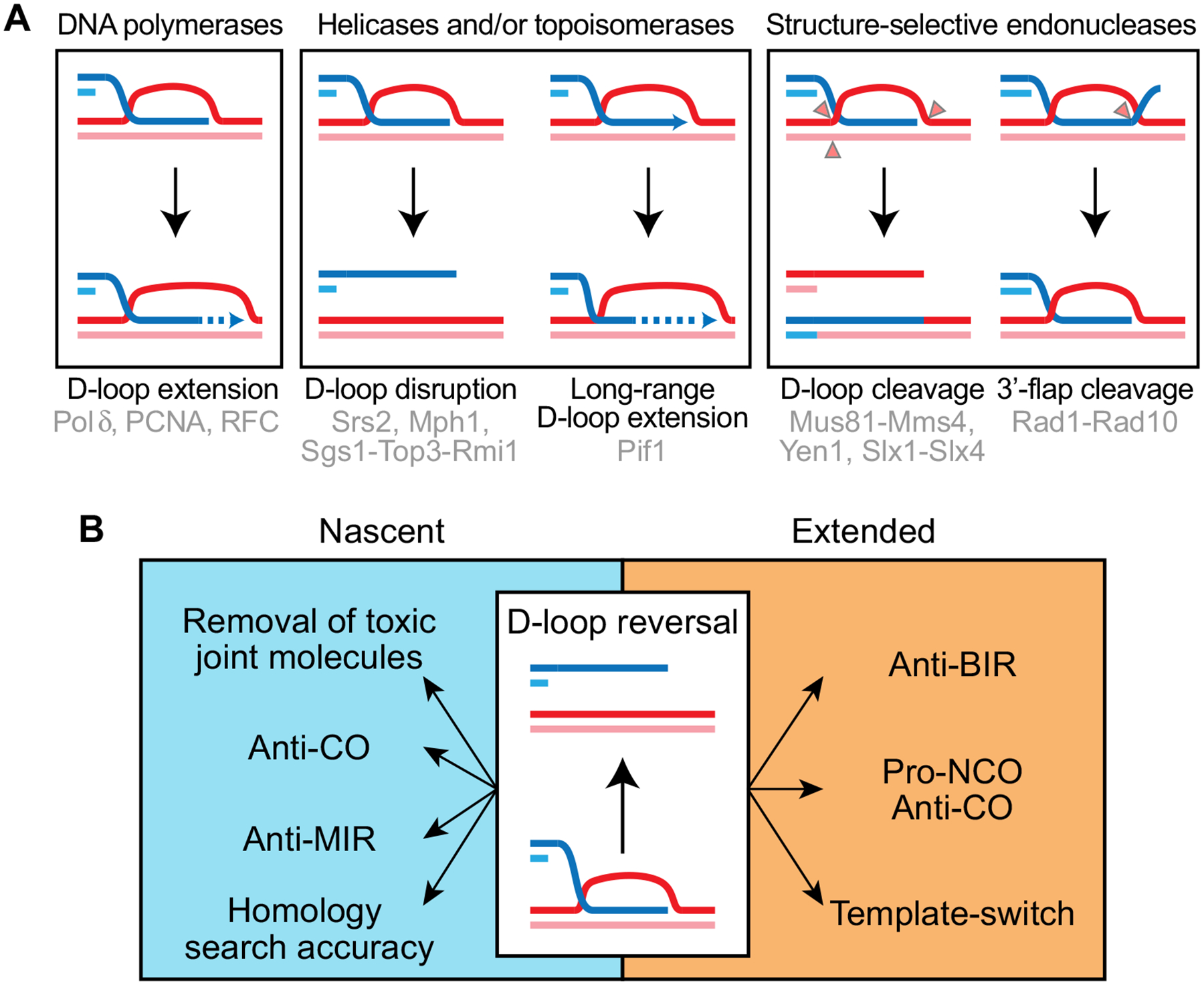
Multiple proteins affect D-loop metabolism, with consequences for HR execution and outcome. a D-loop metabolism is controlled by three main classes of enzymes. b Consequences of nascent and extended D-loop reversal for HR execution, fidelity, and outcome. BIR break-induced replication, MIR multi-invasion induced rearrangement, NCO non-crossover, CO crossover
DNA polymerases that can extend the 3′ extremity of the hDNA and restore the sequence information disrupted by the DSB (McVey et al. 2016).
Helicases and/or topoisomerases that catalyze reversal of nascent and extended D-loops (Crickard and Greene 2019; Heyer 2015) or promote their extension by polymerases (Buzovetsky et al. 2017; Wilson et al. 2013).
Structure-selective endonucleases (SSEs) that can recognize and cleave the strand exchange junctions. This can provide a substrate for DNA polymerase or preclude multi-invasions formation, by cleaving 3′ flap upon internal hDNA formation. Alternatively, nicked strands are substrates for ligases that can covalently join the invading and the invaded dsDNA. Depending on the cleaved strands, this reaction can lead either to the formation of half-crossovers (half-CO) (Deem et al. 2008; Smith et al. 2009) or the formation of a replication fork (Mayle et al. 2015).
The action of SSEs and polymerases represent milestones in HR because they covalently alter the DNA substrate, complicating or preventing reversal of the pathway. Notably, because only few nucleotides suffice for annealing to the opposite end of the DSB, D-loop extension by a DNA polymerase is a tipping point of HR, converting D-loop disruption from a mechanism of anti-recombination to a mechanism of maturation towards the non-crossover (NCO) repair outcome (Fig. 2b). This reaction also inhibits the long-range DNA synthesis required for BIR and stimulates template-switching (Jain et al. 2016b; Luke-Glaser and Luke 2012; Ruiz et al. 2009; Stafa et al. 2014). Consequently, despite being of similar nucleic acids structure, we distinguish nascent D-loop from extended D-loop, and both likely also differ in the types of proteins bound to them.
D-loops exist in a dynamic equilibrium in S. cerevisiae cells
D-loop reversal by various proteins such as the Srs2 and Mph1 3′–5′ helicases, as well as the Sgs1-Top3-Rmi1 (STR) helicasetopoisomerase complex in S. cerevisiae, has long been postulated from genetic studies and physical end-point assays, and supported by biochemical evidence (Coic et al. 2011; Ira et al. 2003; Jain et al. 2016b; Liu et al. 2017; Mazon and Symington 2013; Mitchel et al. 2013; Prakash et al. 2009; Ruiz et al. 2009; Sebesta et al. 2013; Stafa et al. 2014; Sun et al. 2008; Tay et al. 2010; Fasching et al. 2015). However, pinpointing their precise in vivo substrate(s) and defining their interactions has proven challenging in the absence of an assay to physically monitor D-loop intermediates of the pathway. This is in contrast to the double Holliday junction intermediate, which has been demonstrated by 2D gel electrophoresis in meiotic and somatic cells (Bzymek et al. 2010; Schwacha and Kleckner 1995). Surprisingly, in meiotic cells the same method that detects single-end invasions, likely a type of D-loop, fails to detect D-loops in somatic cells (Bzymek et al. 2010). We recently overcome this limitation by developing a method to quantify D-loops and provided physical evidence for active reversal of nascent and extended D-loops in somatic S. cerevisiae cells (Fig. 3) (Piazza et al. 2019).
Fig. 3.
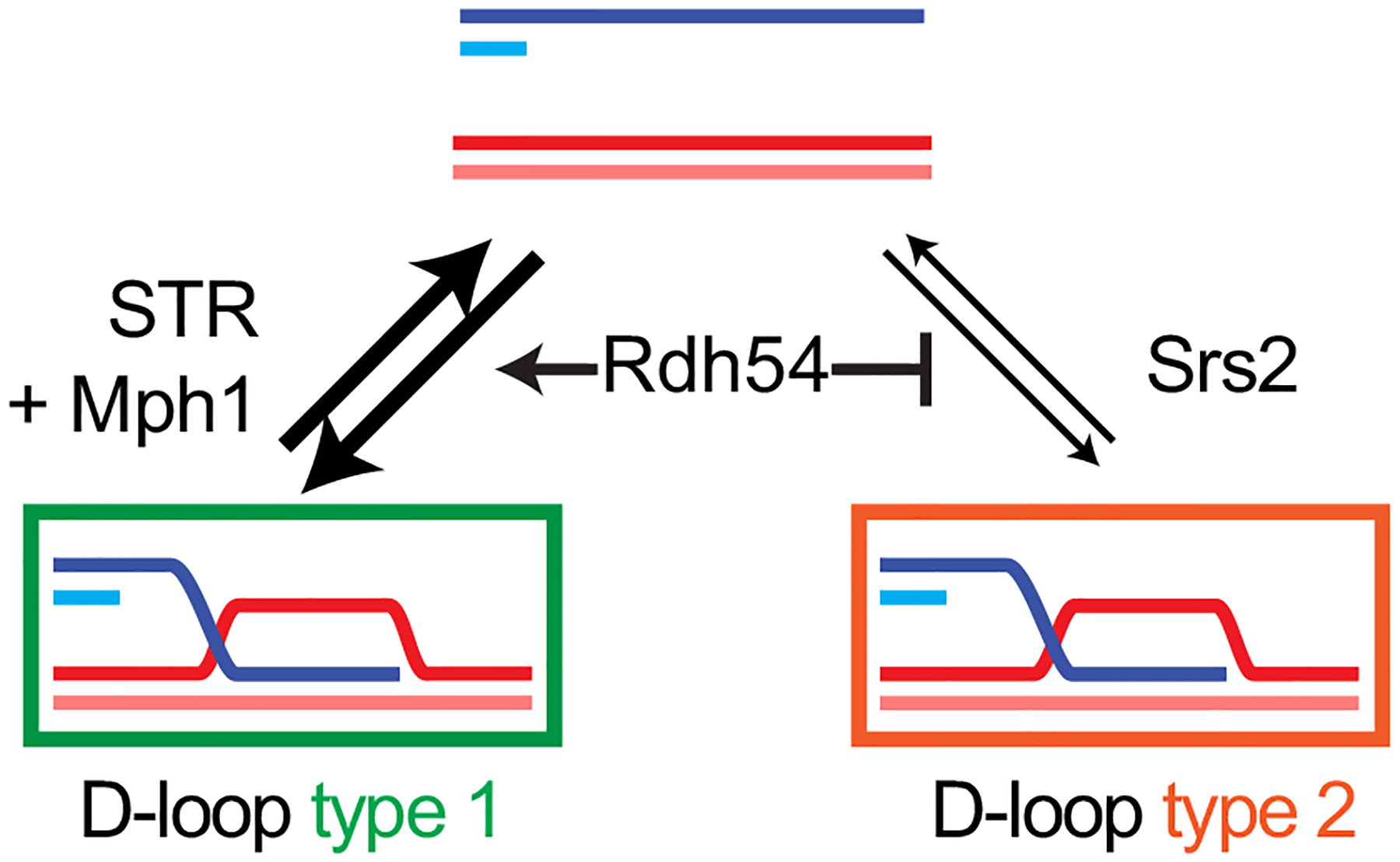
Model for the regulation of nascent D-loop stability in S. cerevisiae. The mechanism by which Rdh54 demarcates these two pathways and/or D-loop types is not known. The human homologs of STR (Sgs1-Top3-Rmi1) is BLM-TOPOIIIα-RMI1/2, of Mph1 is FANCM, and of Rdh54 is possibly RAD54B. Putative functional homologs of Srs2 in human include BLM, FANCJ, FBH1, PARI, RECQ1, RECQ5 and RTEL1. The thickness of the arrows reflects pathway usage
Two types of nascent D-loop are independently targeted by the Srs2 helicase on the one hand, and the Mph1 helicase together with the Sgs1-Top3-Rmi1 (STR) helicasetopoisomerase complex on the other hand (Fig. 3). These two nascent D-loop disruption pathways are delineated by Rdh54 (a Rad54 paralog also known as Tid1) by providing substrates for the Mph1-STR pathway. The nature of these two D-loop types and the mechanism by which Rdh54 distinguishes them remain to be determined. Extended D-loops are mainly disrupted by Mph1 and to a lesser extent by Srs2, while STR appears to be specific for nascent D-loops (Piazza et al. 2019).
Strikingly, these disruption activities targeted the majority of nascent D-loops formed at a 2 kb-long region of perfect homology. Hence, nascent D-loops exist in a dynamic equilibrium in cells, even in the absence of hDNA mismatch recognition (Coic et al. 2011; McVey et al. 2004). It suggests the existence of an unsuspected layer of HR quality control at the nascent D-loop level, which operates in a broader fashion than mismatch-dependent heteroduplex rejection (Hum and Jinks-Robertson 2019; Spies and Fishel 2015). Possible roles for these constitutive D-loop reversal activities include suppressing the formation of half-CO and of multi-invasion-induced rearrangements (MIR) by removing substrates for SSEs (Piazza and Heyer 2019) as well as eliminating toxic joint molecules (Fig. 2b) (Elango et al. 2017; Gangloff et al. 2000; Le Breton et al. 2008). Below, we discuss further potential functions of D-loop reversal in homology search by driving usage of the longest available donor and in suppressing the CO outcome of recombination.
Role of nascent D-loop reversal in homology search
The presence of repeated sequences in the genome can confound the homology search process. This may happen at the level of homology sampling with microhomologies or at short (< 100 bp) repeats (Danilowicz et al. 2015, 2017; Lee et al. 2016; Qi et al. 2015) as well as at the D-loop level as many repeated DNA elements, even in the yeast genome, are larger than the minimal sequence required for efficient HR (Jinks-Robertson et al. 1993; Richard et al. 2008). We suggest that D-loop reversal plays a role in the donor selection process and is thus an integral component of homology search.
First, in the absence of a homologous donor a DSB can be repaired from short (≈ 100 nt) and divergent (≈ 80%) sequences with relatively high efficiency (Anand et al. 2017). This result reveals an astounding mismatch tolerance of homology sampling by the NPF in cells, which operates by microhomology recognition below the length threshold required for sequence uniqueness in bacterial and eukaryotic genomes (Danilowicz et al. 2015, 2017; Lee et al. 2016; Qi et al. 2015). Consequently, the accuracy of homology search not only relies on the intrinsic homology sampling stringency of the NPF but requires also additional regulatory mechanisms.
Second, DNA strand invasion can occur away from the filament terminus in vitro (Adzuma 1992; Piazza et al. 2017; Umlauf et al. 1990; Wright and Heyer 2014). Consistently, sequences located away from the DSB site are efficiently used for HR repair in vivo (Hoang et al. 2010; Inbar and Kupiec 1999; Jain et al. 2016a; Piazza et al. 2017). Hence, in contrast to previous models requiring the DSB to fall into the repeat to initiate ectopic recombination, repeats located in the span of resection are competent for ectopic repair with the risk of causing chromosomal rearrangements (Fig. 4a). This view greatly expands the sequence space in which repeats pose a threat to genomic stability (Fig. 4b) (see Piazza and Heyer 2018 for details). In S. cerevisiae, where the repeated DNA content is relatively low (7%) compared to metazoan and plants, the occurrence of at least one repeat in the resection tract will occur for more than half of DSBs, assuming a resection tract of 8 kb (i.e. equivalent of 2 h of resection; Fig. 4b). Hence, competition of an ectopic homologous match to an internal repeat is presumably a common situation encountered in wild type cells. Constitutive D-loop reversal may limit the use of dispersed repeats, akin to that proposed for RecA synaptic pairing at short (< 100 bp) repeats (Danilowicz et al. 2017). Consistently, D-loop reversal mutants exhibit strongly elevated rates of repeat-mediated chromosomal rearrangements (Argueso et al. 2008; Chan and Kolodner 2011; Hoang et al. 2010; Putnam et al. 2009; Putnam and Kolodner 2017) (reviewed in Piazza and Heyer 2019). This barrier is not absolute and likely involves competition between the dispersed repeats and the allelic donor (or the longest available in the genome).
Fig. 4.
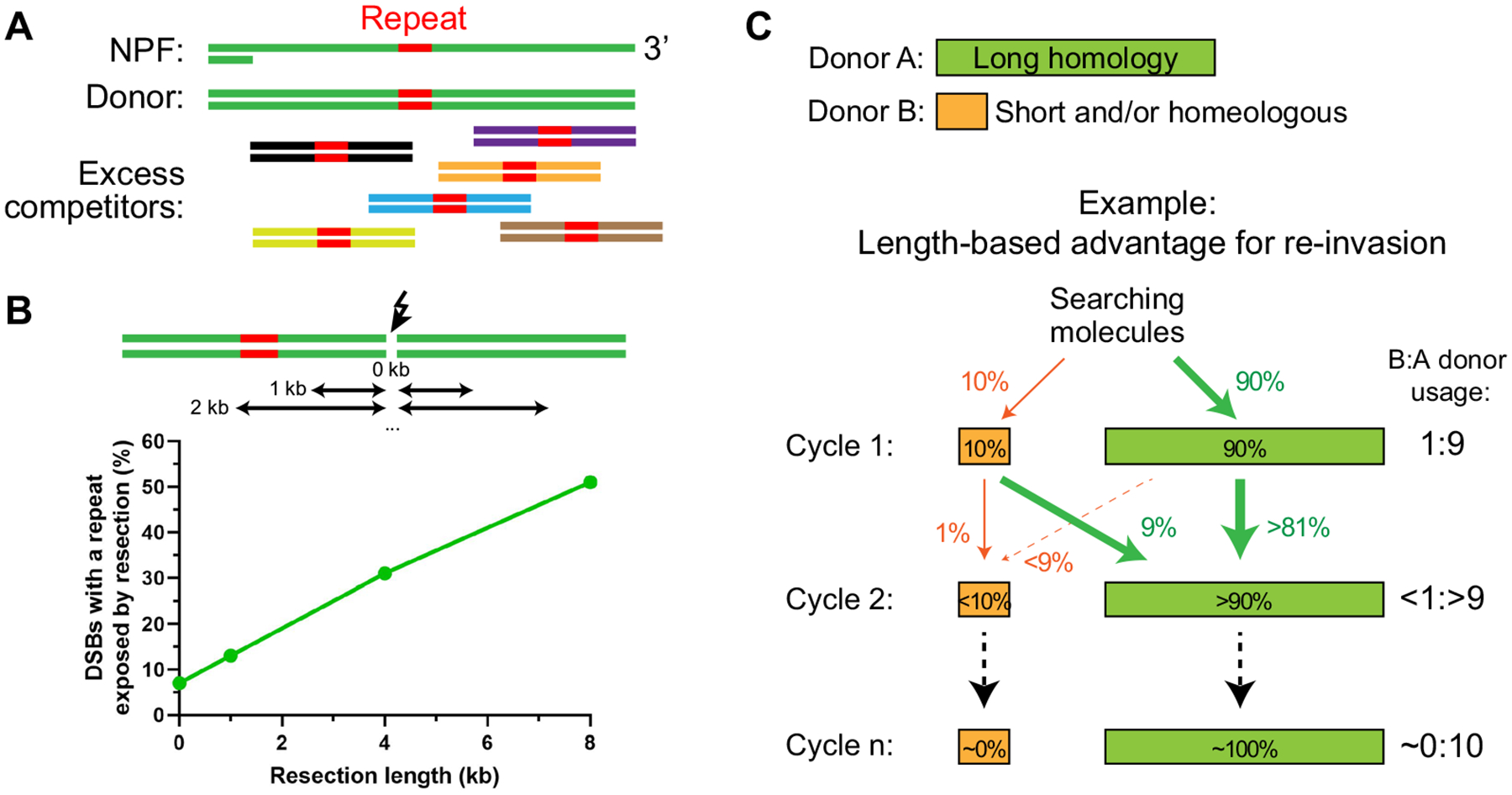
How D-loop reversal can promote repeat bypass during homology search. a Resection can expose DNA sequences that are present in multiple copy in the genome, and efficiently compete against the main (allelic) homologous donor. b Proportion of DSB exposing a repeated region as a function of resection tract length in the S. cerevisiae genome. c Rationale of a kinetic proofreading strategy in which homology length favors (re-)invasion
We propose that reversal of nascent D-loops acts as a second stringency step in the homology search process with the consequence of funneling the searching molecule towards the longest available donor through iterative cycles of recognition/invasion/disruption (Fig. 4c). This seemingly wasteful strategy resembles kinetic proofreading, which increases the specificity of a variety of complex biochemical reactions (Hopfield 1974). It exploits the delay between the binding of two interactors (e.g. the invading strand and the donor) and the initiation of the reaction (e.g. D-loop extension) (Coic et al. 2011). If either the DNA strand re-invasion, the protection against disruption, or the initiation of DNA synthesis is favored by increasing homology length, these cycles will provide bias towards for usage of the longest available homology at each iteration (Fig. 4c). This view broadens the definition of homology search from the sampling by the NPF to include D-loop metabolism up to the initiation of recombination-associated DNA synthesis.
Role of nascent D-loop reversal in promoting a conservative repair outcome
Non-crossover (NCO) is the most conservative HR outcome, because it has no structural or extensive genetic consequences in the subsequent cell generations, contrary to CO, BIR and MIR (Piazza and Heyer 2019). Since the double Holliday Junction (dHJ) intermediate provides opportunity for the formation of CO upon endonucleolytic resolution, avoidance of dHJs represents an anti-CO mechanism. Hence, extended D-loop reversal is both an anti-CO and a pro-NCO reaction by, respectively, directing the pathway away from the dHJ intermediate and towards SDSA. In addition, nascent D-loop reversal may also serve an anti-CO purpose by preventing dual nascent D-loop formation (Fig. 1). Dual nascent D-loops can be directly converted to a dHJ intermediate upon D-loop extension (Fig. 1). They are likely intermediates of the HR pathway owing to the spatial tethering of the two ends of the DSB in somatic cells (Jain et al. 2016a; Lisby et al. 2003), which is expected to favor invasion by the second DSB end upon pioneer invasion by the first end. Hence, limiting the lifetime of nascent D-loops is expected to disproportionately decrease chances for dual nascent D-loops and, hence, disfavor formation of dHJ and subsequent CO. Consistently, nucleo-protein determinants that inhibit ectopic recombination presumably by destabilizing nascent D-loops (such as sequence divergence (Tay et al. 2010; Welz-Voegele and Jinks-Robertson 2008), homology length (Inbar et al. 2000; Jinks-Robertson et al. 1993), and the aforementioned D-loop disruption activities (Chan and Kolodner 2011; Myung et al. 2001; Putnam et al. 2009; Putnam and Kolodner 2017) also inhibit CO.
The existence of dual nascent D-loop might be specifically suppressed during meiosis, in which the CO number per chromosome is tightly regulated. Indeed, contrary to somatic HR, the two DSB ends do not remain associated in meiosis, with one end carrying out the homology search while the other remains localized at the chromosome axis (Brown et al. 2015; Zickler and Kleckner 2015). This sequestration of one of the two DSB ends by cohesin (and its release) might be part of the regulation of the NCO/CO decision, which remains poorly understood (Hong et al. 2013; Kim et al. 2010).
Conclusion
The core steps of HR (i.e. from the NPF assembly onwards up to the initiation of recombinational DNA synthesis) involve transient and mainly non-covalent protein-DNA and DNA-DNA interactions. Reversibility of these associations is likely paramount at multiple steps of the pathway. We propose that nascent D-loop reversibility plays key roles in two aspects of HR fidelity: in homology search by kinetic proofreading and in the regulation of the repair outcome by disfavoring the occurrence of the dHJ intermediate to avoid CO formation.
Acknowledgements
AP was supported by fellowships from the ARC Foundation, the EMBO (ALTF-238-2013), the Framework Project 7 of the European Union (Marie Curie International Outgoing Fellowship 628355) administered by the Institut Pasteur, France, and received financial support from the Philippe Foundation. Research in the WDH laboratory is supported by NIH grants GM58015 and CA92276.
Glossary
- Break-induced replication (BIR)
The long-range DNA synthesis initiated at a DSB site that involves extension of a D-loop
- Crossover (CO) and non-crossover (NCO)
The outcomes of HR repair that, respectively, involves or not the physical exchange of DNA strands between the broken and the donor molecule
- D-loop (or displacement loop)
A DNA joint molecule formed upon pairing of a single-stranded DNA to its complement present in a duplex DNA
- Homologous recombination (HR)
A conserved DSB repair pathway that uses an intact DNA molecule as a template for repair
- Helicase
A protein that uses the energy of ATP hydrolysis to translocate directionally on single-stranded DNA and separate the two strands of duplex DNA
- Multi-invasion-induced rearrangement (MIR)
A tripartite recombination pathway initiated by a DSB end and that causes rearrangement of two independent donor molecules upon endonucleolytic cleavage of D-loops
- Nucleo-protein filament (NPF)
A helical filament formed by the oligomerization of Rad51 (in eukaryotes) and associated proteins onto the single-stranded DNA formed on each side of the break by resection
- Structure-selective endonuclease (SSE)
A protein that recognizes and selectively cleaves DNA structures that are more complex than single-stranded DNA or duplex DNA (such as flaps, D-loops, Holliday junctions, and forks)
Footnotes
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adzuma K (1992) Stable synapsis of homologous DNA molecules mediated by the Escherichia coli RecA protein involves local exchange of DNA strands. Genes Dev 6:1679–1694 [DOI] [PubMed] [Google Scholar]
- Anand R, Beach A, Li K, Haber J (2017) Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Nature 544:377–380. 10.1038/nature22046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso JL, Westmoreland J, Mieczkowski PA, Gawel M, Petes TD, Resnick MA (2008) Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci USA 105:11845–11850. 10.1073/pnas.0804529105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JC, Kowalczykowski SC (2016) RecA: regulation and mechanism of a molecular search engine. Trends Biochem Sci 41:491–507. 10.1016/j.tibs.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordelet H, Dubrana K (2019) Keep moving and stay in a good shape to find your homologous recombination partner. Curr Genet 65:29–39. 10.1007/s00294-018-0873-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Szakal B (2017) Building up and breaking down: mechanisms controlling recombination during replication. Crit Rev Biochem Mol Biol 52:381–394. 10.1080/10409238.2017.1304355 [DOI] [PubMed] [Google Scholar]
- Brown MS, Grubb J, Zhang A, Rust MJ, Bishop DK (2015) Small Rad51 and Dmc1 complexes often co-occupy both ends of a meiotic DNA double strand break. PLoS Genet 11:e1005653 10.1371/journal.pgen.1005653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzovetsky O, Kwon Y, Pham NT, Kim C, Ira G, Sung P, Xiong Y (2017) Role of the Pif1-PCNA complex in Pol delta-dependent strand displacement DNA synthesis and break-induced replication. Cell Rep 21:1707–1714. 10.1016/j.celrep.2017.10.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N (2010) Double Holliday junctions are intermediates of DNA break repair. Nature 464:937–941. 10.1038/nature08868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JE, Kolodner RD (2011) A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet 7:e1002089 10.1371/journal.pgen.1002089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coic E, Martin J, Ryu T, Tay SY, Kondev J, Haber JE (2011) Dynamics of homology searching during gene conversion in Saccharomyces cerevisiae revealed by donor competition. Genetics 189:1225–1233. 10.1534/genetics.111.132738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickard JB, Greene EC (2019) Helicase mechanisms during homologous recombination in Saccharomyces cerevisiae. Annu Rev Biophys. 10.1146/annurev-biophys-052118-115418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilowicz C, Yang D, Kelley C, Prevost C, Prentiss M (2015) The poor homology stringency in the heteroduplex allows strand exchange to incorporate desirable mismatches without sacrificing recognition in vivo. Nucleic Acids Res 43:6473–6485. 10.1093/nar/gkv610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilowicz C, Hermans L, Coljee V, Prevost C, Prentiss M (2017) ATP hydrolysis provides functions that promote rejection of pairings between different copies of long repeated sequences. Nucleic Acids Res 45:8448–8462. 10.1093/nar/gkx582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Barker K, VanHulle K, Downing B, Vayl A, Malkova A (2008) Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 179:1845–1860. 10.1534/genetics.108.087940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango R, Sheng Z, Jackson J, DeCata J, Ibrahim Y, Pham NT, Liang DH, Sakofsky CJ, Vindigni A, Lobachev KS, Ira G, Malkova A (2017) Break-induced replication promotes formation of lethal joint molecules dissolved by Srs2. Nat Commun 8:1790 10.1038/s41467-017-01987-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching CL, Cejka P, Kowalczykowski SC, Heyer WD (2015) Top3-Rmi1 dissolve Rad510-mediated D loops by a topoisomerase-based mechanism. Mol Cell 57:595–606. 10.1016/j.molcel.2015.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet 25:192–194 [DOI] [PubMed] [Google Scholar]
- Heyer WD (2015) Regulation of recombination and genomic maintenance. Cold Spring Harb Perspect Biol 7:a016501 10.1101/cshperspect.a016501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang ML, Tan FJ, Lai DC, Celniker SE, Hoskins RA, Dunham MJ, Zheng YX, Koshland D (2010) Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet 6:e1001228 10.1371/journal.pgen.1001228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Sung YJ, Yu M, Lee M, Kleckner N, Kim KP (2013) The logic and mechanism of homologous recombination partner choice. Mol Cell 51:440–453. 10.1016/j.molcel.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield JJ (1974) Kinetic proofreadig: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA 71:4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hum YF, Jinks-Robertson S (2019) Mismatch recognition and subsequent processing have distinct effects on mitotic recombination intermediates and outcomes in yeast. Nucleic Acids Res. 10.1093/nar/gkz126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar O, Kupiec M (1999) Homology search and choice of homologous partner during mitotic recombination. Mol Cell Biol 19:4134–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar O, Liefshitz B, Bitan G, Kupiec M (2000) The relationship between homology length and crossing over during the repair of a broken chromosome. J Biol Chem 275:30833–30838. 10.1074/jbc.C000133200 [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Sugawara N, Haber JE (2016a) Role of double-strand break end-tethering during gene conversion in Saccharomyces cerevisiae. PLoS Genet 12:e1005976 10.1371/journal.pgen.1005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Sugawara N, Mehta A, Ryu T, Haber JE (2016b) Sgs1 and Mph1 helicases enforce the recombination execution checkpoint during DNA double-strand break repair in Saccharomyces cerevisiae. Genetics 203:667–675. 10.1534/genetics.115.184317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S, Michelitch M, Ramcharan S (1993) Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol 13:3937–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang LR, Jordan A, Dekker J, Kleckner N (2010) Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143:924–937. 10.1016/j.cell.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton C, Dupaigne P, Robert T, Le Cam E, Gangloff S, Fabre F, Veaute X (2008) Srs2 removes deadly recombination intermediates independently of its interaction with SUMO-modified PCNA. Nucleic Acids Res 36:4964–4974. 10.1093/nar/gkn441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Qi Z, Greene EC (2016) ATP hydrolysis promotes duplex DNA release by the RecA presynaptic complex. J Biol Chem 291:22218–22230. 10.1074/jbc.M116.740563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R (2003) Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol 5:572–577 [DOI] [PubMed] [Google Scholar]
- Liu J, Ede C, Wright WD, Gore SK, Jenkins SS, Freudenthal BD, Todd Washington M, Veaute X, Heyer WD (2017) Srs2 promotes synthesis-dependent strand annealing by disrupting DNA polymerase delta-extending D-loops. Elife. 10.7554/eLife.22195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S, Luke B (2012) The Mph1 helicase can promote telomere uncapping and premature senescence in budding yeast. PLoS One 7:e42028 10.1371/journal.pone.0042028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, Bjergbaek L, Lupski JR, Ira G (2015) Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science 349:742–747. 10.1126/science.aaa8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon G, Symington LS (2013) Mph1 and Mus81-Mms4 prevent aberrant processing of mitotic recombination imtermediates. Mol Cell 52:63–74. 10.1016/j.molcel.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Adams M, Staeva-Vieira E, Sekelsky JJ (2004) Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167:699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Khodaverdian VY, Cerqueira P, Heyer W-D (2016) Eukaryotic DNA polymerases in homologous recombination. Annu Rev Genet 50:393–421. 10.1146/annurevgenet-120215-035243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Haber JE (2014) Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol 6:a016428 10.1101/cshperspect.a016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman Z, Lottersberger F, Takai H, Kibe T, Gong Y, Takai K, Bianchi A, Zimmermann M, Durocher D, de Lange T (2018) 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature 560:112–116. 10.1038/s41586-018-0324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K, Lehner K, Jinks-Robertson S (2013) Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet 9:e1003340 10.1371/journal.pgen.1003340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Chen C, Kolodner RD (2001) SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet 27:113–116 [DOI] [PubMed] [Google Scholar]
- Piazza A, Heyer WD (2018) Multi-invasion-induced rearrangements as a pathway for physiological and pathological recombination. BioEssays 40:e1700249 10.1002/bies.201700249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Heyer WD (2019) Homologous recombination and the formation of complex genomic rearrangements. Trends Cell Biol 29:135–149. 10.1016/j.tcb.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Wright WD, Heyer WD (2017) Multi-invasions are recombination byproducts that induce chromosomal rearrangements. Cell 170(760–773):e715 10.1016/j.cell.2017.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Shah SS, Wright WD, Gore SK, Koszul R, Heyer WD (2019) Dynamic processing of displacement loops during recombinetional DNA repair. Mol Cell 73(1255–1266):e1254 10.1016/j.molcel.2019.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, Ira G (2009) Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev 23:67–79. 10.1101/gad.1737809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Kolodner RD (2017) Pathways and mechanisms that prevent genome instability in Saccharomyces cerevisiae. Genetics 206:1187–1225. 10.1534/genetics.112.145805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hayes TK, Kolodner RD (2009) Specific pathways prevent duplication-mediated genome rearrangements. Nature 460:984–989. 10.1038/nature08217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Redding S, Lee JY, Gibb B, Kwon Y, Niu HY, Gaines WA, Sung P, Greene EC (2015) DNA sequence alignment by microhomology sampling during homologous recombination. Cell 160:856–869. 10.1016/j.cell.2015.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Dujon B (2008) Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev 72:686–727. 10.1128/MMBR.00011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JF, Gomez-Gonzalez B, Aguilera A (2009) Chromosomal trans-locations caused by either Pol32-dependent or Pol32-independent triparental break-induced replication. Mol Cell Biol 29:5441–5454. 10.1128/mcb.00256-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N (1995) Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83:783–791 [DOI] [PubMed] [Google Scholar]
- Sebesta M, Burkovics P, Juhasz S, Zhang SF, Szabo JE, Lee M, Haracska L, Krejci L (2013) Role of PCNA and TLS polymerases in D-loop extension during homologous recombination in humans. DNA Repair 12:691–698. 10.1016/j.dnarep.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Lam AF, Symington LS (2009) aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol Cell Biol 29:1432–1441. 10.1128/mcb.01469-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M, Fishel R (2015) Mismatch repair during homologous and homeologous recombination. Cold Spring Harb Perspect Biol 7:a022657 10.1101/cshperspect.a022657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafa A, Donnianni RA, Timashev LA, Lam AF, Symington LS (2014) Template switching during break-induced replication is promoted by the Mph1 helicase in Saccharomyces cerevisiae. Genetics 196:1017–1028. 10.1534/genetics.114.162297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC (2008) The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell 32:118–128. 10.1016/j.molcel.2008.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS (2016) Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol 51:195–212. 10.3109/10409238.2016.1172552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay YD, Sidebotham JM, Wu L (2010) Mph1 requires mismatch repair-independent and -dependent functions of MutS alpha to regulate crossover formation during homologous recombination repair. Nucleic Acids Res 38:1889–1901. 10.1093/nar/gkp1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf SW, Cox MM, Inman RB (1990) Triple-helical DNA pairing intermediates formed by recA protein. J Biol Chem 265:16898–16912 [PubMed] [Google Scholar]
- Welz-Voegele C, Jinks-Robertson S (2008) Sequence divergence impedes crossover more than noncrossover events during mitotic gap repair in yeast. Genetics 179:1251–1262. 10.1534/genetics.108.090233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, Ira G (2013) Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 502:393–396. 10.1038/nature12585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WD, Heyer WD (2014) Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D-loop formation. Mol Cell 53:420–432. 10.1016/j.molcel.2013.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WD, Shah SS, Heyer WD (2018) Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem 293:10524–10535. 10.1074/jbc.TM118.000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N (2015) Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harbor Perspect Biol 7:a016626 10.1101/cshperspect.a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Fabre E (2019) Chromatin mobility upon DNA damage: state of the art and remaining questions. Curr Genet 65:1–9. 10.1007/s00294-018-0852-6 [DOI] [PubMed] [Google Scholar]


