Figure 1.
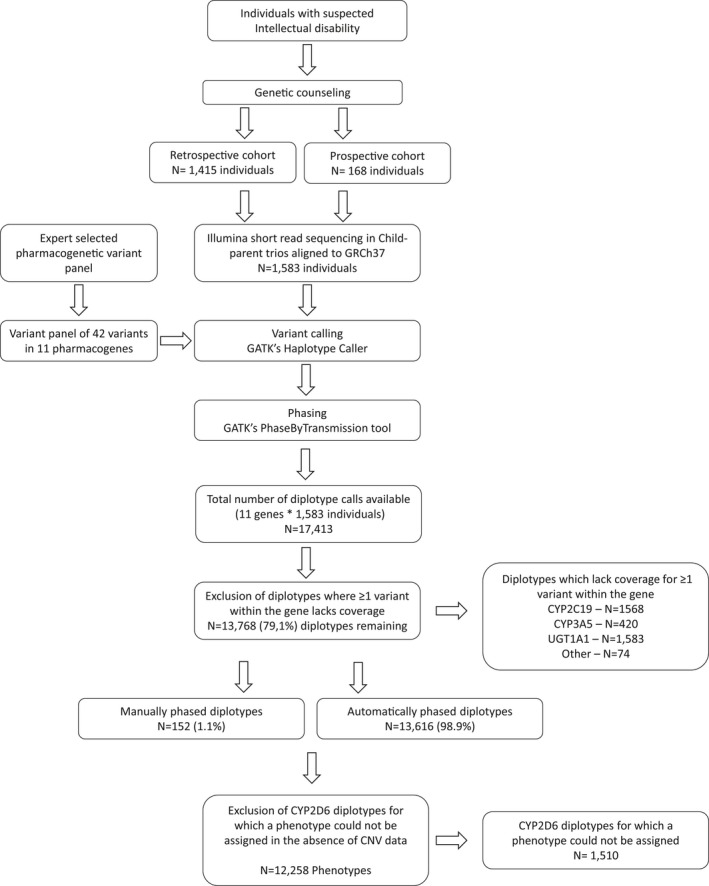
Study flowchart. Whole exome sequencing data from individuals sequenced for diagnostics was used to obtain a clinically relevant pharmacogenetics (PGx) profile. Retrospective cohort: individuals sequenced prior to August 2016; prospective cohort: individuals sequenced after August 2016 if they opted in for obtaining their PGx profile. The expert selected PGx panel was obtained from the Ubiquitous Pharmacogenomics U‐PGx consortium. Sufficient coverage was classified as haplotype quality of at least 20. Due to the absence of copy number variants (CNVs), only CYP2D6 diplotypes consisting of two null‐alleles were included as CNVs would not change the phenotype assignment. Manual phasing and phenotype assignments were based on translation tables from the U‐PGx consortium. GATK, Genome Analysis Tool Kit.
