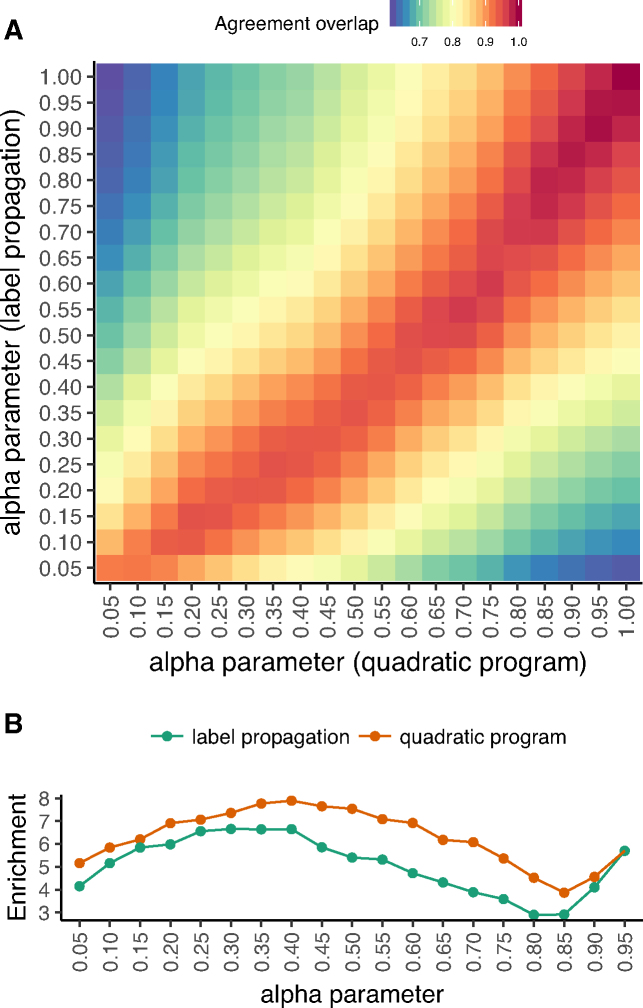
Figure 4.
Quadratic programming and label propagation adsorption approaches yield similar jointly determined specificities. We consider the similarity of results between our two distinct approaches to sharing DNA binding information across structurally similar proteins. (A) For different values of α for the QP approach (x-axis) and for the LPA approach (y-axis), we plot the Jaccard coefficient of the sets of corresponding columns in agreement across PW-2015 and NM-2015 (i.e. the agreement overlap). Lower overlap is indicated by blue and higher overlap is indicated by red. (B) As a function of α (x-axis), we compare the agreement gain versus agreement loss ratio (i.e., as described in Figure 2; y-axis) when using either the LPA approach (green) or the QP approach (red, also shown in Figure 2, bottom). The QP approach obtains higher ratios for all α and the ratio peaks at 0.3  α
α  0.4 for both approaches.
0.4 for both approaches.