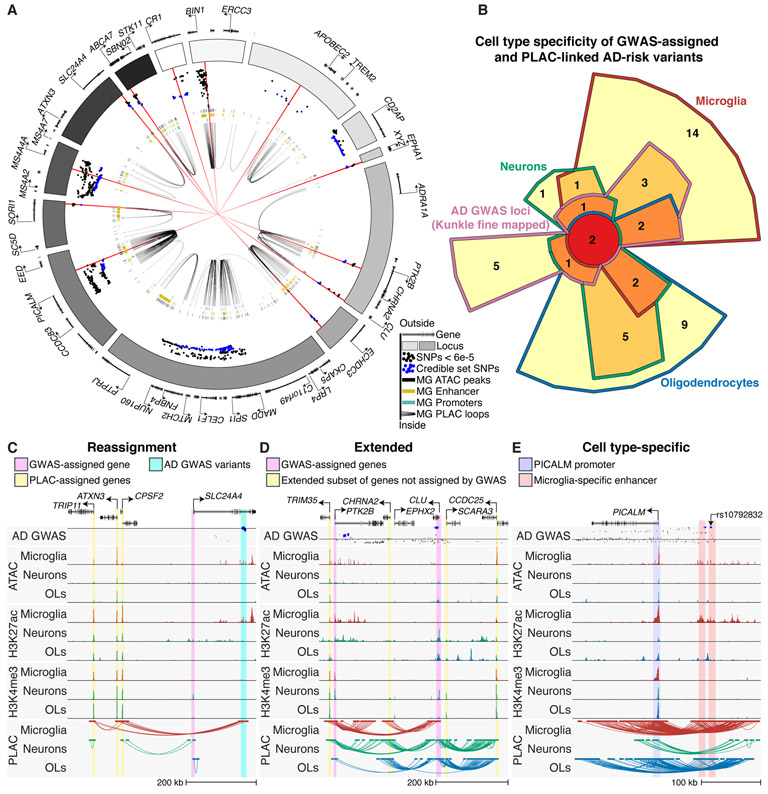
Fig. 3.
Expanded gene network of AD-risk loci. (A) Circos plot of AD GWAS loci, showing microglia enhancers (gold bars), promoters (turquoise), open chromatin regions (black bars) and PLAC-seq interactions (black loops). Dots show z-score values of high-confidence AD variants identified by fine mapping (Kunkle, stage 1) with log10 p-value < 6e-5 (18). Blue dots represent z-score values of the credible set of AD SNPs (95% confidence); red lines show 15 high-confidence AD SNPs with a posterior probability > 0.2. (B) Chow-Ruskey plot of genes that are GWAS-assigned and PLAC-seq linked to AD-risk credible set variants in microglia, neurons and oligodendrocytes. (C)-(E) UCSC browser of interactions at AD-risk loci demonstrating (C) reassignment of GWAS-assigned genes, (D) extension of GWAS-assigned genes and (E) cell type-specific gene regulatory regions. The AD GWAS track shows meta-analysis p-values of stage 2 variants (18); line indicates p-value = 5e-8; blue dots are fine mapped 95% credible set variants.