SUMMARY
Canonical plant phytochromes are master regulators of photomorphogenesis and the shade avoidance response. They are also part of a widespread superfamily of photoreceptors with diverse spectral and biochemical properties. Plant phytochromes belong to a clade including other phytochromes from glaucophyte, prasinophyte, and streptophyte algae (all members of the Archaeplastida) and those from cryptophyte algae. This is consistent with recent analyses supporting existence of an AC (Archaeplastida + Cryptista) clade. AC phytochromes have been proposed to arise from ancestral cyanobacterial genes via endosymbiotic gene transfer (EGT), but most recent studies instead support multiple horizontal gene transfer (HGT) events to generate extant eukaryotic phytochromes. In principle, this scenario would be compared to the emerging understanding of early events in eukaryotic evolution to generate a coherent picture. Unfortunately, there is currently a major discrepancy between the evolution of phytochromes and the evolution of eukaryotes; phytochrome evolution is thus not a solved problem. We therefore examine phytochrome evolution in a broader context. Within this context, we can identify three important themes in phytochrome evolution: deletion, duplication, and diversification. These themes drive phytochrome evolution as organisms evolve in response to environmental challenges.
Keywords: endosymbiosis, evolution, light harvesting, photosynthesis, shade avoidance
I. INTRODUCTION
Modern agriculture relies on planting crop species at high density to maximize yield for a given amount of arable land (Fedoroff et al., 2010). However, high-density planting means that individual plants can shade each other, triggering competition (Casal, 2013). Plants detect such competition via light quality and react by derepression of the shade avoidance response, with important agricultural consequences (Casal, 2013; Ballaré & Pierik, 2017). In 1959, the photoreceptor controlling this process was identified as phytochrome (Butler et al., 1959). Phytochrome detects the presence of competition by measuring the ratio of photosynthetically active red light to photosynthetically inactive far-red light, a ratio that varies due to harvesting of red light by the competitor (Fig. 1). Phytochrome also functions as a master regulator of photomorphogenesis to control germination, flowering, and almost everything in between. Thousands of genes are activated or repressed in the model plant species Arabidopsis thaliana in response to phytochrome signaling (Rockwell et al., 2006; Hu et al., 2009; Franklin & Quail, 2010; Strasser et al., 2010; Chen & Chory, 2011; Hu et al., 2013), so phytochrome research remains important both for plant biology and for photobiology in general. Signal transduction by phytochrome is a complex field in its own right, but it is clear that activation of plant phytochromes by red light triggers translocation to the nucleus (Nagatani, 2004).
Figure 1. Phytochrome and shade sensing.
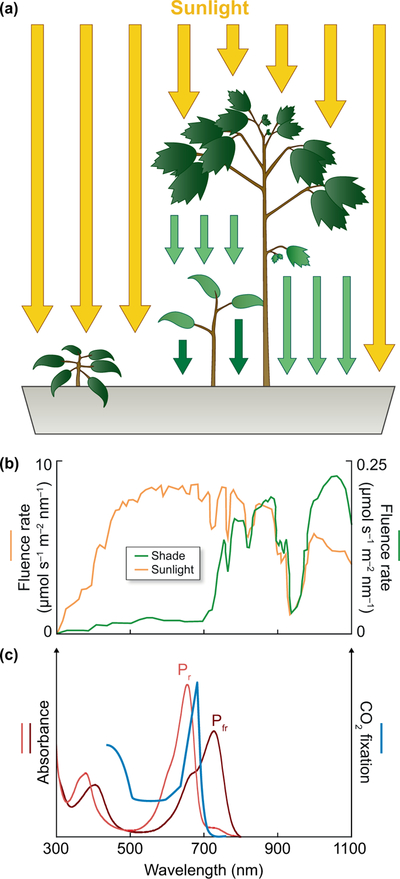
(a) Plants growing in direct sunlight or under shade from competitors are exposed to different light environments, leading to distinct morphology and pigment development. The plant on the left is in direct sunlight, but the lower plant on the right is shaded by a competing plant of a different species. Scattering is omitted for clarity. (b) A competing canopy depletes light for shaded plants. In the case of tropical rain forests, attenuation is approximately 40-fold for the near-infrared and greater for photosynthetically useful red light. Units are μmol s−1 m−2 nm−1 for both axes. The red:far-red ratio (R:FR) is 1.25 in direct sunlight and 0.31 in shade (Lee & Graham, 1986). (c) The action spectrum for carbon fixation in the green alga Hydrodictyon is compared to absorption spectra for the red-absorbing Pr and far-red-absorbing Pfr photostates of wheat phytochrome C (Raven, 1969; Chen et al., 2014).
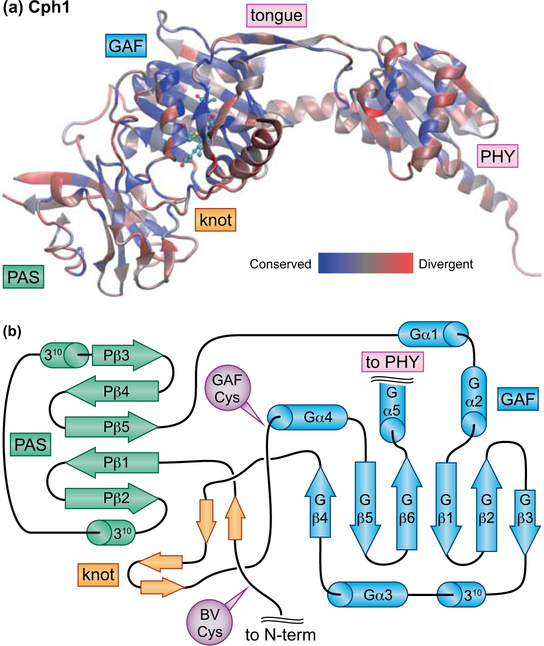
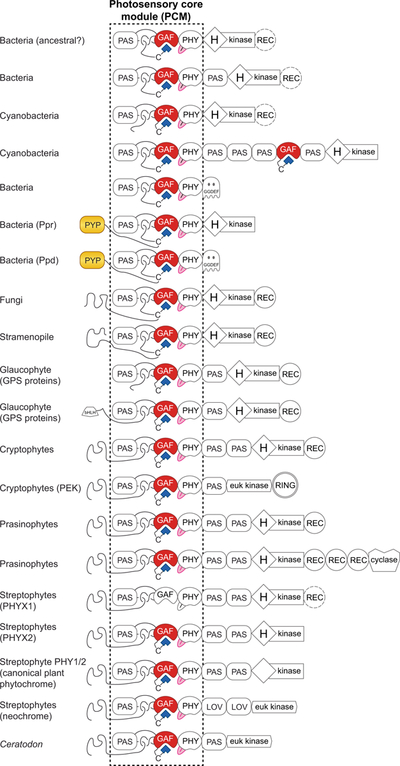
Canonical plant phytochromes are large (ca. 1100 amino acids) photoreceptors that have a conserved domain architecture including a three-domain photosensory core module (PCM, Fig. 2) near the N-terminus. The PCM has PAS (Fig. 2), GAF, and PHY domains that adopt a conserved knotted structure (Wagner et al., 2005; Essen et al., 2008; Yang et al., 2008; Burgie et al., 2014a). In this knot (Fig. 2B), an N-terminal extension upstream of the PAS domain passes through a loop internal to the GAF fold (Essen et al., 2008; Yang et al., 2008). Light perception by phytochrome requires a linear tetrapyrrole (bilin) chromophore derived from heme, and plant phytochromes adopt a red-absorbing C5–Z,syn C10–Z,syn C15–Z,anti configuration in the dark-adapted state (Pr, Fig. 1C). Light absorption triggers a 15,16–photoisomerization reaction that generates a far-red-absorbing C5–Z,syn C10–Z,syn C15–E,anti photoproduct (Pfr, Fig. 1C) which can regenerate Pr upon light absorption. 15,16–photoisomerization underlies the signature red/far-red reversibility of phytochrome action in plant biology. Pfr can also decay to Pr in the absence of light (dark or thermal reversion). The stability of Pfr is thus determined by the light environment, the presence of possible binding partners that modulate the rate of dark reversion, the widely varying rate of dark reversion among different phytochromes, and the temperature at which the light-independent reaction is occurring. Most phytochromes follow this pattern, but bathyphytochromes instead adopt the 15E Pfr form as the dark-stable state with a 15Z photoproduct (Rockwell et al., 2006). Typically dimeric, phytochromes are found in plants (canonical plant phytochromes), bacteria, fungi, and diverse eukaryotic algae (Fig. 3A). These diverse phytochromes share the knotted PCM fused to different C-termini in different organisms (see below).
Figure 2. Structure of the knotted phytochrome PCM.
(a) Cyanobacterial phytochrome Cph1 from Synechocystis sp. PCC 6803 (Essen et al., 2008) is shown color-coded by sequence similarity, calculated using a published sequence alignment (Rockwell et al., 2006) and the BLOSUM62 matrix as implemented in homolmapper (Rockwell & Lagarias, 2007). (b) Topology cartoon for the knotted PAS-GAF region of the PCM with the knot in orange, the PAS domain in green, and the GAF domain in blue. Cys residues used for chromophore ligation are indicated in purple. Secondary structure elements are numbered from the N-terminus of each domain for the PAS and GAF domains, with the domain indicated by P and G, respectively. 1-turn 310 helices and short β-strands in the knot are indicated but not numbered. Domain names: GAF, cGMP phosphodiesterase/Adenylate cyclase/FhlA; PAS, Per/ARNT/Sim; PHY, phytochrome-specific. BV, biliverdin; N-term, N-terminus; tongue, conserved insertion loop specific to the PHY domain.
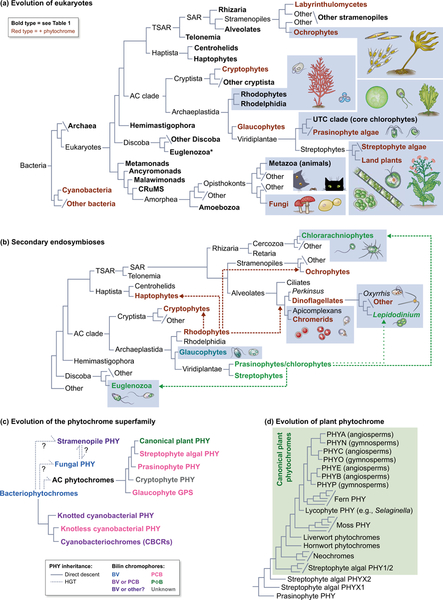
Figure 3. Phytochrome and the evolution of eukaryotes.
(a) A simplified working view of the tree of life is shown, drawing on recent studies (Duanmu et al., 2014; Wickett et al., 2014; Burki et al., 2016; Derelle et al., 2016; Brown et al., 2018; Lax et al., 2018; Gawryluk et al., 2019; Strassert et al., 2019). Bold type indicates taxa assessed for phytochrome and bilin biosynthesis in Table 1. Lineages containing at least some organisms in which phytochromes are present are indicated in red. Representative organisms are shown for selected lineages (not to scale; enclosed in dotted boxes). Branch lengths are arbitrary. Asterisk, see legend to Table 1 for possible phytochromes in Euglenozoa. CRuMS, supergroup containing collodictyonids, rigifilids, and Mantamonas; AC clade, Archaeplastida + Cryptista; UTC clade, Ulvophyceae, Trebouxiophyceae and Chlorophyceae; TSAR, Telonemia + SAR; SAR, supergroup containing Stramenopiles, Alveolates, and Rhizaria. (b) Secondary endosymbioses in eukaryotes. Acquisition of photosynthesis in cryptophytes, haptophytes, ochrophytes, and photosynthetic alveolates (chromerids and dinoflagellates) occurred via secondary endosymbiosis of a rhodophyte (dashed red lines). Acquisition of photosynthesis in euglenozoa and chlorarachniophytes occurred via secondary endosymbiosis of prasinophyte or chlorophyte algae (green dashed lines). “Green dinoflagellates” of the genus Lepidodinium have replaced the ancestral rhodophyte-derived plastid via secondary endosymbiosis of a prasinophyte or chlorophyte alga (green dotted lines). Representative organisms are shown for selected lineages as in (a). Colors indicate plastid lineage; nonphotosynthetic plastids are not colored. (c) A schematic view of phytochrome evolution is shown. Colors indicate chromophore precursors incorporated by phytochromes from different taxa: BV only, blue; BV or PCB, purple; BV or unknown bilin, deep purple (Rockwell et al., 2014); PCB, pink; PΦB, green; unknown, grey. Dashed lines indicate inferred HGT events. PHY, phytochrome; BV, biliverdin IXα; PCB, phycocyanobilin; PΦB, phytochromobilin. (d) A schematic view of plant phytochrome evolution is shown, demonstrating that canonical plant phytochromes are part of a larger clade of streptophyte phytochromes using the nomenclature of (Li et al., 2015).
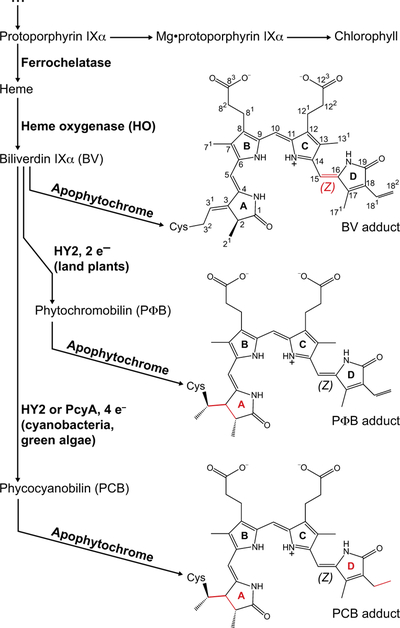
Photoconversion thus requires a bilin with a 15,16–double bond (Murphy & Lagarias, 1997). Three suitable chromophore precursors have been reported to date (Fig 4). Canonical plant phytochromes utilize phytochromobilin, or PΦB, as the chromophore precursor (Lagarias & Rapoport, 1980; Rockwell et al., 2006). PΦB is autocatalytically attached to a conserved Cys residue in the GAF domain (the GAF Cys: Fig. 2B). Phytochromes from streptophyte algae, sister to land plants, also have this Cys residue but instead synthesize phycocyanobilin, or PCB (Wu et al., 1997; Rockwell et al., 2017). Both of these phycobilins are synthesized from heme via two steps (Fig. 4). Heme oxygenase (HO) first converts heme to biliverdin IXα (BV, Fig. 4). In the second step, the ferredoxin-dependent bilin reductase (FDBR) HY2 converts BV into PCB or PΦB (Frankenberg et al., 2001; Kohchi et al., 2001; Rockwell et al., 2017). PΦB synthesis from BV is a 2-electron reduction, whereas PCB synthesis is a 4-electron reduction (Fig. 4). PCB is also used by some bacterial phytochromes (Yeh et al., 1997; Jaubert et al., 2007), including cyanobacterial ones, but these organisms use the FDBR PcyA rather than HY2. FDBRs are ubiquitous in photosynthetic eukaryotes (Rockwell & Lagarias, 2017), with other family members (PEBA and PEBB) producing phycoerythrobilin for use in bilin-based light-harvesting systems (Glazer, 1985; Frankenberg et al., 2001). Eukaryotic PCYA enzymes are found in glaucophyte, prasinophyte, and chlorophyte algae, defined as in (Duanmu et al., 2014), as well as some rhodophytes and cryptophytes (see below). HY2 is currently only known from streptophytes (Fig. 3A). Eukaryotic FDBRs are plastid localized (Kohchi et al., 2001; Duanmu et al., 2013), and it is not known how bilin moves to the cytosol for assembly with apophytochrome. This process could be even more complicated in algae which acquired photosynthesis via secondary endosymbiosis (Fig. 3B).
Figure 4. Biosynthesis of bilin chromophores.
The pathway from heme to biliverdin IXα (BV) and reduced phycobilins (PCB and PΦB) is shown. Free bilins are then assembled with apophytochrome to yield covalent adducts, examples of which are shown on the right in the C5–Z,syn, C10–Z,syn, C15–Z,anti configuration (Wagner et al., 2005; Essen et al., 2008; Burgie et al., 2014a). The numbering system is indicated for BV. For simplicity, a single structure is shown; however, other BV adducts have been observed (Salewski et al., 2013). For BV the photoactive 15,16–double bond is highlighted in red. For PCB and PΦB, sites of reduction relative to BV are highlighted in red. The degree of reduction relative to BV is indicated (e–, electron).
However, not all phytochromes require reduced phycobilins synthesized by FDBRs. Phytochromes from nonphotosynthetic and anoxygenic phototrophic bacteria incorporate BV as chromophore precursor (Bhoo et al., 2001; Giraud et al., 2002), requiring only HO for chromophore biosynthesis. Such proteins are often designated bacteriophytochromes. These proteins retain the knotted PAS-GAF-PHY PCM, but BV is covalently attached to a conserved Cys residue upstream of the knot (the BV Cys: Fig. 2B). This Cys is absent in canonical plant phytochromes, but some phytochromes from cyanobacteria and ochrophytes (photosynthetic stramenopiles) have both Cys residues. The authentic chromophore composition of such cases is currently unknown. Distantly related cyanobacteriochrome photoreceptors (CBCRs, see below) typically contain the conserved GAF Cys residue and incorporate PCB as initial chromophore precursor (Ishizuka et al., 2011; Rockwell et al., 2012a), but exceptional cases using a different Cys residue for attachment (Fushimi et al., 2016) or ligating BV to the GAF Cys (Narikawa et al., 2015; Fushimi et al., 2019) are also known. BV is also incorporated as chromophore by fungal phytochromes and by some phytochromes from cyanobacteria and ochrophytes (Blumenstein et al., 2005; Froehlich et al., 2005; Quest et al., 2007; Fortunato et al., 2016). Thus, there is no known case in which phytochrome incorporates a chromophore other than BV or a reduced phycobilin derived from BV. The photosensory function of phytochrome thus requires oxygen for synthesis of BV.
Photoconversion triggers structural changes in the dimeric configuration of phytochrome and refolding of a conserved “tongue” loop in the PHY domain (Fig. 2A) that contacts the chromophore-binding pocket in the GAF domain (Burgie et al., 2014b; Takala et al., 2014; Burgie et al., 2016). These structural responses to photoconversion provide different exposed surfaces for protein-protein interaction that appear to be generally conserved in knotted phytochromes and to modulate activity of C-terminal “output” domains. Plant phytochromes translocate from the cytosol to the nucleus upon photoconversion (Nagatani, 2004), where they modulate gene expression by interacting with phytochrome-interacting factors (PIFs), a family of basic helix-loop-helix (bHLH) transcription factors (Franklin & Quail, 2010; Chen & Chory, 2011; Pham et al., 2018). Upon interaction with phytochrome, PIFs are phosphorylated and targeted for degradation by the proteasome in a complex process involving multiple protein kinases, multiple ubiquitin ligases, the COP1/SPA complex, and other factors (Pham et al., 2018).
Light-dependent nuclear accumulation of phytochrome has also been observed in the prasinophyte alga Micromonas pusilla (Duanmu et al., 2014), so it seems likely that at least some aspects of phytochrome signal transduction are conserved in Viridiplantae (prasinophytes + streptophytes, Fig. 3A) and perhaps in other eukaryotes. However, there are cyanobacterial members of the phytochrome superfamily that lack the knotted 3-domain PCM (see below). We can thus define a superfamily of phytochrome photoreceptors by the presence of a bilin-binding GAF domain, with three families: knotted phytochromes (PAS-GAF-PHY), knotless phytochromes (GAF-PHY), and CBCRs (GAF-only). CBCRs do not have a PHY domain and consequently exhibit different structural changes upon photoconversion (Burgie et al., 2013; Narikawa et al., 2013; Lim et al., 2014; Lim et al., 2018).
The importance of canonical plant phytochromes for agriculture and the diversity of the phytochrome superfamily beg the question of phytochrome evolution. Where did phytochrome originate? How and when did plants acquire phytochrome? What drives the evolution of phytochrome? Recent studies have shown that the evolution of phytochrome is a complex process, and it is not currently possible to reconcile recent studies of phytochrome evolution with our understanding of eukaryotic evolution. We will summarize the present state of affairs, highlight areas of ongoing uncertainty, and attempt to place phytochrome evolution within the larger contexts of phytochrome photobiology and eukaryotic evolution. We argue that phytochrome has crossed from bacteria to eukaryotes more than once, with one of these transfers giving rise to modern plant phytochromes and with none of those transfers originating within cyanobacteria. We identify three themes driving phytochrome evolution: deletion, duplication, and diversification.
II. PHYTOCHROME ORIGINS: IT ALL STARTS IN BACTERIA
Both PAS and GAF domains are found in other proteins, and the PHY domain also adopts a GAF fold (Essen et al., 2008; Yang et al., 2008). However, it is not currently possible to identify a clear sister lineage to the phytochrome GAF domain. In the absence of an outgroup, we can consider the prerequisites for the rise of functional phytochrome photoreceptors. Light perception and signal transduction by known phytochromes requires a bilin chromophore that can undergo 15,16–photoisomerization (Murphy & Lagarias, 1997) and hence requires oxygen and HO.
Are there ancient alternatives? Alternative chromophores include heme, heme precursors such as porphyrin, and phyllobilins derived from chlorophyll breakdown (Kräutler, 2014). Phyllobilins seem to be a later innovation in photosynthetic organisms, but ancestral phytochromes evolved in cells containing heme and porphyrins. Phytochrome variants are known to bind porphyrins in recombinant expression systems (Fischer et al., 2005; Wagner et al., 2008), so there may have been an early stage in phytochrome evolution in which phytochromes bound porphyrins but did not carry out photoisomerization. It may also be possible to make suitable bilins in the absence of oxygen. Parallel oxygen-dependent and oxygen-independent pathways are known for the synthesis of heme (Dailey et al., 2017). Alternative oxygen-dependent heme breakdown pathways have also been discovered (Matsui et al., 2013; Nambu et al., 2013), as has an oxygen-independent pathway (LaMattina et al., 2016). The oxygen-independent pathway is not widespread, implying a recent origin, and the resulting anaerobilin product is thought to lack a 15,16–double bond. Nevertheless, this finding raises the possibility that suitable bilins could be made by cells in the absence of oxygen.
In speculating on the origins of phytochrome, it is also important to consider phytochrome in the absence of bilin. There are known functions for canonical plant phytochromes in the cytosol prior to photoconversion (Paik et al., 2012; Hughes, 2013), so similar functions could have been carried out by apophytochrome or by phytochrome with bound porphyrin. Phytochrome function in the absence of HO has also been reported in the anoxygenic photosynthetic bacterium Rhodopseudomonas palustris (Fixen et al., 2014), so either the apoprotein is biologically functional in this system or there is a labile alternative chromophore that was lost during purification. R. palustris also has an unusual phytochrome that has apparently lost light sensing altogether and functions as a redox-sensing histidine kinase, regulating kinase activity via redox-sensitive disulfide bonds (Vuillet et al., 2007). We speculate that early ancestors of phytochromes could have bound different tetrapyrroles or had some other sensing function. Unfortunately, we have no evidence for oxygen-independent BV synthesis. Similarly, the achromatic phytochrome of R. palustris is proposed to have evolved from photosensory ancestors (Vuillet et al., 2007), so it is not a reliable basis for inferring an ancestral achromatic phytochrome.
We thus cannot reliably infer the features of a phytochrome before the advent of HOs. We therefore begin with a photosensory phytochrome having a knotted PCM and a bound BV chromophore; in other words, we postulate a founder phytochrome resembling a modern bacteriophytochrome. One can instead argue that it would be simpler to begin with an isolated bilin-binding GAF domain similar to a modern CBCR. However, knotless phytochromes and CBCRs are only known from cyanobacteria. Moreover, the knotted PCM is much more widespread and therefore seems more plausible as the ancestral state. BV-containing phytochromes also are more widespread in both prokaryotes and eukaryotes (Table 1), and BV binding is less widespread in CBCRs than in knotted phytochromes (Narikawa et al., 2015). BV itself can be made from ubiquitous heme with a single, widespread enzyme, whereas FDBRs are largely confined to photosynthetic organisms (Table 1).
Table 1:
Distribution of Phytochromes and Bilins Across the Tree of Life1
| Lineage | Examples | PHY | HO | FDBR | bilins |
|---|---|---|---|---|---|
| Archaea | Pyrococcus, Lokiarchaeota | − | − | − | none known |
| cyanobacteria | Prochlorococcus, Nostoc | ± | + | + | BV, PCB, other |
| other bacteria | Rhodopseudomonas, Escherichia coli | ± | ± | ± | ±BV, ±PCB |
| metamonads | Giardia, Trichomonas | − | − | − | none known |
| ancyromonads | Fabomonas, Nutomonas | − | fragments2 | − | ±BV? |
| malawimonads3 | Malawimonas, Gefionella | − | − | − | none known |
| CRuMs | Rigifila, Collodictyon | − | − | − | none known |
| Amoebozoa | Paramoeba, Dictyostelium | − | ± | − | ±BV |
| Fungi | yeast, molds, mushrooms | ± | ± | − | ±BV |
| Metazoa | Homo sapiens, birds, bees (animals) | − | ± | − | ±BV |
| Euglenozoa | Euglena, Eutreptiella | fragments4 | + | + | BV, PCB |
| other Discoba | Leishmania, Trypanosoma | − | − | − | none known |
| Hemimastigophora | Hemimastix, Spironema | − | − | − | none known |
| cryptophytes | Guillardia, Hemiselmis | + | + | + | BV, PCB, other |
| other Cryptista | Roombia, Palpitomonas | − | ± | − | ±BV |
| rhodophytes | Galdieria, laver, Cyanidioschyzon | − | + | + | BV, ±PCB, ±other |
| Rhodelphidia | Rhodelphis | − | − | − | none known |
| glaucophytes | Cyanophora | + | + | + | BV, PCB |
| prasinophyte algae | Micromonas, Ostreococcus | ± | + | + | BV, PCB, ±other |
| UTC clade | Chlamydomonas, Volvox | − | + | + | BV, PCB |
| streptophyte algae | Mesostigma, Chara, Klebsormidium | + | + | + | BV, PCB, other |
| land plants | ferns, cacti, redwood trees, bananas | + | + | + | BV, PΦB, ±other |
| centrohelids | Acanthocystis (centroheliozoans) | − | ± | − | ±BV |
| haptophytes | Pavlova, Emiliana, Phaeocystis | − | + | + | BV, ? |
| Telonemia3 | Telonema | fragments | − | − | BV? |
| Rhizaria | Paulinella, foraminiferans | − | ± | ± | ±BV, ±PCB, ±? |
| alveolates | dinoflagellates, ciliates, Plasmodium | − | ± | ± | ±BV, ±PCB, ±? |
| labyrinthulomycetes (slime nets) | Aplanochytrium, QPX pathogen | ± | ± | − | ±BV |
| ochrophytes | diatoms, kelp | ± | + | + | BV, ? |
| other stramenopiles | Cafeteria, Phytophthora | − | ± | − | ±BV |
Drawn from (Duanmu et al., 2014; Keeling et al., 2014; Li et al., 2015; Brown et al., 2018; Lax et al., 2018; Gawryluk et al., 2019; Johnson et al., 2019; Strassert et al., 2019). PHY, phytochrome. See Fig. 3A for an overview of eukaryotic evolution. Bilin composition is listed for cases where a phytochrome chromophore is known or can be inferred from the presence of PCYA or HY2. “Other,” reduced bilins other than PCB or PΦB are known to be present. “?,” FDBRs are present, but their reaction products are unknown. Fragments: short fragments are present in one or more transcriptomes. Such cases have not been phylogenetically analyzed.
Short sequences closely related to heme oxygenases are present in the deposited raw sequence reads form the transcriptome of Fabomonas tropica NYK3C (SRA SRX3153020 in the Genbank Sequence Read Archive), so this organism may contain a heme oxygenase.
Raw sequence reads for Gefionella okellyi strain 249 and Telonema isolate P-1 are available from the Genbank Sequence Read Archive (SRA SRX3152400 and SRA SRR7371268, respectively). Short sequences closely related to heme oxygenases are present in the latter, so Telonema may contain a heme oxygenase. However, it should be noted that the gene content of other members of these lineages may vary.
Possible phytochrome sequences are present in one Eutreptiella transcriptome but have not been phylogenetically analyzed; similar sequences are not found in other genomes and transcriptomes from Euglenozoa. The provenance of these sequences is thus unclear.
We therefore view evolution of diverse PCMs having the GAF Cys, lacking the knot and PAS domain, and lacking both PAS and PHY domains as subsequent events. Known pathways for BV synthesis require HO and hence would have evolved after oxygen levels were high enough. Heme and porphyrin can promote formation of reactive oxygen species in the presence of light and oxygen (photosensitization), and HO is more widespread than phytochrome in both prokaryotes and eukaryotes (Table 1). HO may thus have provided a means for living cells to suppress this toxic combination. HO generates BV, carbon monoxide, and free iron as reaction products, permitting iron scavenging (Ratliff et al., 2001; Fujii et al., 2004; Gisk et al., 2012; Duanmu et al., 2013). Coupling BV to a photoreceptor would provide an elegant means for bacteria to use a single pathway for both sensing and detoxifying the toxic combination of light and oxygen.
What were these bacteria? The use of light-harvesting phycobiliproteins by cyanobacteria (Glazer, 1985) also requires HO and FDBRs; hence, cyanobacteria would possess the requisite chromophore precursors. However, recent phylogenetic analyses recover knotted cyanobacterial phytochromes as a derived clade within other bacterial phytochromes (Duanmu et al., 2014; Li et al., 2015). Moreover, heterotrophic bacteria would also find it advantageous to avoid the toxic cocktail of light, oxygen, and tetrapyrroles. Such organisms might well use HO for detoxification under hyperoxia or redox stress, like modern mammals, and hence would also be suitable candidates. We still must acknowledge the important caveat that an ancestral cyanobacterial lineage of phytochromes may have been completely supplanted by extant examples, but current evidence implicates heterotrophic or anoxygenic phototrophic bacteria as the origin of phytochrome because of the close relationships between their phytochromes and knotted cyanobacterial phytochromes (Duanmu et al., 2014; Li et al., 2015). Such bacteria lack FDBRs, and many possess phytochrome operons comprised of heme oxygenase (BphO) and bacteriophytochrome (BphP) genes (Auldridge & Forest, 2011). These operons could have provided a selective advantage by enabling adaptive responses to diurnal changes in light and oxygen levels as their cyanobacterial neighbors carried out photosynthesis. We envisage that phytochromes were subsequently co-opted by cyanobacteria, either for similar responses or for regulation of the newly emerging family of light-harvesting phycobiliproteins. Phycobiliproteins greatly expanded the wavelength range for cyanobacterial photosynthesis (Glazer, 1985), but they are metabolically expensive and hence could have been regulated by phytochrome early in cyanobacterial evolution in the same way that modern regulation of phycobiliproteins is carried out by CBCRs (see below).
Under these assumptions, the PCM would have subsequently transferred to eukaryotes. Canonical plant phytochromes are part of a large clade (Fig. 3C) also including phytochromes from prasinophyte, cryptophyte, and glaucophyte algae (Duanmu et al., 2014; Li et al., 2015; Kooss & Lamparter, 2017); hereafter, we designate these as AC phytochromes for Archaeplastida + Cryptista (Burki et al., 2016). Phytochromes from ochrophytes and fungi have also been characterized (Blumenstein et al., 2005; Froehlich et al., 2005; Rockwell et al., 2014; Fortunato et al., 2016). More recent transcriptomic studies (Keeling et al., 2014; Johnson et al., 2019) have revealed the presence of phytochromes in labyrinthulomycetes (a nonphotosynthetic stramenopile lineage, Fig. 3A), raising the possibility that phytochrome was present in stramenopiles before acquisition of photosynthesis in the ochrophyte clade (Derelle et al., 2016). Current studies do not support a single lineage of eukaryotic phytochromes (Duanmu et al., 2014; Li et al., 2015). Instead, there is a robust AC phytochrome clade and a possible phylogenetic relationship between ochrophyte and fungal phytochromes (Duanmu et al., 2014), but there is no association between AC phytochromes and other eukaryotic phytochromes (Fig. 3C). It has been proposed that cyanobacterial sequences are sister to AC phytochromes (Kooss & Lamparter, 2017), implying that AC phytochromes are derived from cyanobacterial ancestors via endosymbiotic gene transfer (EGT). However, analyses with more extensive sampling instead place knotted cyanobacterial phytochromes as a derived, independent clade (Duanmu et al., 2014; Li et al., 2015). Current studies do not rule out the possibility of genetic drift in extant cyanobacterial sequences or the possibility that an earlier cyanobacterial phytochrome lineage has been supplanted in modern prokaryotes. However, under the assumption of an ancestral bacteriophytochrome, both current statistically robust studies indicate that phytochromes would have transferred into eukaryotes at least twice: once to give rise to the AC phytochrome lineage and at least once more to give rise to other eukaryotic phytochromes (Fig. 3C).
Within the AC clade, glaucophyte phytochromes are the first lineage to diverge from other AC phytochromes (Fig. 3C), followed by cryptophyte phytochromes. Prasinophyte and streptophyte phytochromes form a clade, as do these organisms (Duanmu et al., 2014; Wickett et al., 2014; Li et al., 2015). However, the placement of cryptophyte phytochromes does not match the placement of these algae within the AC clade (Fig. 3A and C). Early evolution of the AC clade is not fully understood (Burki et al., 2016; Strassert et al., 2019). Recent studies provide evidence that rhodophytes diverged first within Archaeplastida, with glaucophytes as sister to Viridiplantae (Lax et al., 2018; Price et al., 2019). This result has gained further support with the very recent discovery of the Rhodelphidia, nonphotosynthetic flagellates sister to rhodophytes (Gawryluk et al., 2019). Cryptophytes thus would be derived members of the Cryptista (sister to Archaeplastida within the AC clade, Fig. 3A), having acquired photosynthesis via secondary endosymbiosis of a rhodophyte (Fig. 3B). However, this pattern is in fundamental conflict with the evolution of AC phytochromes: cryptophyte phytochromes are recovered as sister to those from Viridiplantae (Fig. 3C). Future studies will be needed to resolve this discrepancy. However, our current level of knowledge is sufficient to identify three themes that play a major part in the evolution of phytochrome: deletion, duplication, and diversification.
III. DELETION: LIFE WITHOUT PHYTOCHROME
Tracing the evolution of phytochromes in eukaryotes presents challenges similar to that of doing so for other proteins potentially acquired from prokaryotes. Unless the protein in question is ubiquitous in eukaryotes, it is not straightforward to distinguish between multiple introductions to eukaryotes and a single introduction with subsequent losses. Invoking deletions minimizes the number of transfers to eukaryotes that would be required to explain the distribution of eukaryotic phytochromes; thus, one could envision a single introduction of PHY into eukaryotes with multiple subsequent losses. Might phytochrome have been present in the ancestral eukaryote? The possible relationship between stramenopile and fungal PHY could support such a model but could also be explained by horizontal gene transfer (HGT). Indeed, streptophyte phytochromes provide an example of HGT: one lineage of land plant phytochromes, the neochromes, i transferred from bryophytes to ferns and then within ferns via HGT (Li et al., 2014). Neochromes are chimeric phytochrome-phototropin fusions retaining the phytochrome PCM that are confined to hornworts, ferns, and streptophyte algae of the Zygnemataceae. All neochrome lineages descended from a single ancestral PCM, but the algal and hornwort/fern C-termini have distinct origins (Li et al., 2015). This may implicate additional HGT events. Therefore, the barrier preventing multiple, independent transfers of phytochrome from bacteria to eukaryotes via HGT may be intellectual rather than evolutionary.
However, this reasoning does not preclude loss of phytochrome during evolution. A simple analysis of the distribution of phytochrome in recent transcriptomes and genomes make it clear that phytochrome loss does occur. Phytochromes from diatoms, phaeophyte algae (kelps or brown algae), and giant viruses infecting phaeophytes (Schroeder et al., 2009) form a clade (Duanmu et al., 2014; Li et al., 2015), implicating a single ancestral phytochrome in ochrophytes. Phytochromes are present in the diatoms Thalassiosira antarctica, T. miniscula, T. pseudonana, T. rotula, and T. weissflogii but are absent in T. oceanica (Armbrust et al., 2004; Lommer et al., 2012; Johnson et al., 2019). This situation could arise due to multiple HGT events after initial introduction into ochrophytes, but as many as four would be required in Thalassiosira alone. It thus seems less complex to imagine a middle ground combining fewer HGT events with subsequent loss of phytochrome in T. oceanica and other cases.
Could phytochrome loss resolve the conundrum posed by cryptophyte phytochromes? It is clear that rhodophytes have undergone substantial genome reduction (Collén et al., 2013; Qiu et al., 2017), so the absence of phytochromes in extant rhodophytes may be part of this process. Indeed, there is evidence for loss of FDBRs during evolution of rhodophyte and cryptophyte algae: polyextremophilic Cyanidioschyzon spp. contain only PCYA (Rockwell & Lagarias, 2017; Rossoni et al., 2019), whereas many other rhodophytes instead contain PEBA and PEBB (Rockwell & Lagarias, 2017). Cryptophytes have rhodophyte-derived secondary plastids (Fig. 3B), and most cryptophytes contain only PEBA and PEBB. Some cryptophytes also contain PCYA derived from Cyanidioschyzon PCYA (Rockwell & Lagarias, 2017), but such cryptophytes do not form a clade. Therefore, either a subset of mesophilic cryptophytes acquired PCYA from Cyanidioschyzon via two or more HGT events or all three FDBRs were present in the ancestor to the cryptophyte plastid. The latter scenario implies independent loss of one or more of these enzymes from rhodophytes and cryptophytes after establishment of secondary endosymbiosis in cryptophytes. A similar process would provide an admittedly speculative explanation for the close relationship between cryptophyte phytochromes and those from Viridiplantae, but in this case one would assume that ancestral rhodophyte phytochromes were transferred to the cryptophyte nucleus via EGT but were lost in extant rhodophytes.
Even so, the distribution of phytochromes in extant eukaryotes is not consistent with an ancestral eukaryotic phytochrome. Eukaryotic phytochromes are confined to three major eukaryotic assemblages (MEA): stramenopiles belong to the SAR or TSAR supergroup (Strassert et al., 2019), Archaeplastida and Cryptista belong to the AC supergroup (Burki et al., 2016), and Fungi belong to the Amorphea (Fig. 3A). Moreover, phytochrome-containing taxa are in clades that are well nested within Amorphea, AC, and SAR/TSAR (Fig. 3A). Ancient HGT among those lineages could explain the possible relationship between fungal and stramenopile phytochromes (Duanmu et al., 2014), but AC phytochromes are a distinct lineage. We therefore conclude that deletion of phytochromes is a frequent occurrence during evolution, but that this does not support an ancestral eukaryotic phytochrome. Instead, phytochrome has crossed from bacteria into eukaryotes at least twice. After these transfers, there have been deletion events in some lineages, but there have also been duplications.
IV. DUPLICATION: MANY PHYTOCHROMES FROM ONE
Even though many organisms lack phytochrome altogether, others have large numbers of phytochrome photoreceptors. Arabidopsis and related mustards have five phytochromes, but one of these (PHYD) is a late duplication found only in the flowering plant family Brassicaceae (Mathews, 2006; Mathews & McBreen, 2008). The model kelp Ectocarpus siliculosus has four phytochromes (Cock et al., 2010), as do some Hemiselmis strains (Keeling et al., 2014; Johnson et al., 2019). Remarkably, phytochrome gene families have apparently undergone much greater expansion in glaucophytes, with as many as eleven members of this family in Cyanophora paradoxa (Price et al., 2019). Such duplication is not limited to eukaryotes; indeed, R. palustris has six phytochromes, serving as sensors for light and redox conditions (Giraud et al., 2002; Giraud et al., 2005; Vuillet et al., 2007; Fixen et al., 2014). Cyanobacteria can contain a lavish panoply of photoreceptors, as shown by the more than twenty members of the phytochrome superfamily found within the genome of Nostoc punctiforme (Rockwell et al., 2015a). Hence, duplication of phytochrome genes is clearly an important factor in phytochrome evolution.
What can duplication tell us about the evolution of canonical plant phytochromes? Remarkably, multiple duplication events occurred during evolution of plant phytochrome itself (Fig. 3D). In a recent study (Li et al., 2015), Mathews and colleagues discovered unexpectedly high levels of phytochrome diversity in the streptophyte algal clades that are the closest relatives of land plants (Mesostigmatales, Klebsormidiales, Desmidiales, Coleochaetales, and Zygnematales). Land plant phytochromes are members of the streptophyte PHY1/2 lineage (Fig. 3D), and all members of this lineage share the canonical plant phytochrome domain architecture (see below). Within this clade, the gene phylogenies generally match that of the organisms (Wickett et al., 2014), although there is evidence of early duplications and subsequent losses (Li et al., 2015). Remarkably, two additional phytochrome lineages were found outside this lineage but within the larger clade of streptophyte phytochromes. These are the PHYX1 and PHYX2 genes, which apparently occur only in the Coleochaetales and Zygnematales (Fig. 3D). These organisms are more derived orders of streptophyte algae (Wickett et al., 2014), so the placement of PHYX1 and PHYX2 is at odds with the organismal phylogeny. This discrepancy provides further evidence of gene duplication and subsequent loss. Canonical plant phytochromes thus arose via duplication in streptophyte algae that also contained PHYX1 and PHYX2, with subsequent deletion of PHYX1 and PHYX2 in most streptophytes (Li et al., 2015). PHYX1 and PHYX2 proteins have a different C-terminal architecture than those of canonical plant phytochromes (see below), so we speculate that the master regulator function of plant phytochromes arose with or after the duplication event establishing the canonical plant phytochrome lineage. Subsequent duplications in land plants (Fig. 3D) have given rise to ‘light-stable’ PHYN/A and ‘light-labile’ PHYP/B/D/E canonical plant phytochrome lineages (Mathews, 2006; Li et al., 2014; Li et al., 2015), allowing the generation of a range of responses to different light environments and illustrating the link between duplication and diversification.
V. DIVERSIFICATION: A CONSERVED PCM IN DIFFERENT CONTEXTS
In most phytochromes, the PCM is fused to variable C-termini, with an N-terminal PCM that could simplify formation of the knot. Prokaryotic phytochromes do not have large N-terminal extensions, with the noteworthy exception of hybrid Ppr and Ppd photosensors from some purple bacteria (Kyndt et al., 2005; Kyndt et al., 2007; Kyndt et al., 2010). In these proteins, an N-terminal PYP (photoactive yellow protein) domain precedes the PCM (Fig. 5); it is not clear whether the knot forms in this case, but the PYP domain is known to affect the photosensory properties of the PCM (Kyndt et al., 2007). Many eukaryotic phytochromes have longer extensions N-terminal to the knot (Fig. 5). For example, streptophyte and prasinophyte phytochromes have a Ser/Thr rich region upstream of the knot that possesses sites for autophosphorylation and inter-domain contacts (Rockwell & Lagarias, 2006; Han et al., 2010a; Velazquez Escobar et al., 2017). This region is thought to be unstructured and can be removed without affecting photoconversion or knot formation, but it is required for proper function in vivo (Uenaka & Kadota, 2007; Han et al., 2010b). Fungal and stramenopile phytochromes have even longer extensions of unknown function which similarly lack recognizable structural elements (Rockwell et al., 2006). Glaucophytes contain phytochromes with shorter N-termini similar to those of prokaryotic phytochromes, but they also contain uncharacterized sequences with longer N-terminal extensions that show sequence similarity to basic helix-turn-helix DNA-binding domains (Price et al., 2019). Thus, regions of phytochrome that lack universal conservation or recognizable domains have diversified and potentially acquired distinct biological functions.
Figure 5. Diverse domain architectures in knotted phytochromes.
Jellybean domain diagrams are shown for diverse prokaryotic and eukaryotic phytochromes that use the canonical knotted PCM (Duanmu et al., 2014; Li et al., 2015). Bilin-binding GAF domains are red, the tongue region of the PHY domain is pink, bilin chromophores are shown as blue polygons, and chromophore-binding PYP domains are yellow. REC domains that are present in only some cases are dashed, as in PHYX1. Domain names: GAF, cGMP phosphodiesterase/Adenylate cyclase/FhlA; PAS, Per/ARNT/Sim; PHY, phytochrome-specific; (H)kinase, histidine kinase bidomain, with the presence of the His indicated by H; REC, response regulator receiver; RING, really interesting new gene; euk kinase, eukaryotic protein kinase; cyclase, eukaryotic adenylate/guanylate cyclase; GGDEF, diguanylate cyclase; PYP, photoactive yellow protein; bHLH, basic helix-loop-helix; LOV, PAS domain belonging to the light/oxygen/voltage lineage. Two-Cys phytochromes from cyanobacteria and glaucophytes are not indicated.
C-terminal to the PCM, phytochromes from different organisms have different domain architectures. Bacterial phytochromes typically have a PCM associated with a functional histidine kinase bidomain (i.e., containing the phosphoacceptor His residue), as in the well-characterized model phytochrome Cph1 from Synechocystis sp. PCC 6803 (Hughes et al., 1997; Yeh et al., 1997). Some examples, such as Anacy_3347 from the cyanobacterium Anabaena cylindrica PCC 7122, also have a C-terminal response regulator (REC) domain (Fig. 5). Others, such as EPX62760 from the myxobacterium Cystobacter fuscus DSM 2262 and ADC61659 from the purple bacterium Allochromatium vinosum DSM 180T, have a PAS domain between the PHY domain and the His kinase module, again with or without a C-terminal REC domain (Fig. 5). In another variation, the genome of the cyanobacterium Microcoleus sp. PCC7113 encodes a large protein with an N-terminal knotted PCM and C-terminal histidine kinase bidomain linked by multiple PAS domains and a single CBCR (Fig. 5). There are also examples linking the phytochrome PCM to other bacterial signal transduction pathways; e.g., the α-proteobacteria, R. palustris and Bradyrhizobium sp. strain ORS278, contain phytochromes with C-terminal S-boxes implicated in protein-protein interactions (Giraud et al., 2002). Moreover, some phytochromes and hybrid Ppd photoreceptors have a C-terminal GGDEF domain (Fig. 5) that can synthesize the bacterial second messenger cyclic-di-GMP (Kyndt et al., 2005; Ryu & Gomelsky, 2014).
C-terminal domain architectures also vary in eukaryotic phytochromes. Stramenopile and fungal phytochromes have functional histidine kinase bidomains at the C-terminus, typically followed by C-terminal REC domains (Fig. 5). Glaucophyte GPS (Glaucophyte phytochrome sensor) proteins have a single PAS domain followed by a functional histidine kinase bidomain, sometimes with a C-terminal REC domain (Duanmu et al., 2014). Cryptophyte phytochromes are associated with two different termini (Fig. 5). In one of these, the PCM is followed by a single PAS domain, a eukaryotic protein kinase domain, and a RING domain (putative E3 ubiquitin ligase). Such proteins have been termed PEKs for phytochrome eukaryotic kinase (Duanmu et al., 2014), but little is known about their properties and functions. In other cryptophyte phytochromes, the PCM is followed by a pair of PAS domains followed by a histidine kinase bidomain and a REC domain. This architecture is also shared by prasinophyte phytochromes, and the phosphoacceptor His is present in both (Li et al., 2015). Some prasinophyte phytochromes have three REC domains and a possible nucleotide cyclase domain after the histidine kinase module (Fig. 5). One such protein, DtenPHY1 from Dolichomastix tenuilepis CCMP3274, was nevertheless shown to exhibit light-regulated autophosphorylation of the His residue as a recombinantly expressed truncation (Duanmu et al., 2014).
The domain architecture shared by conventional cryptophyte and prasinophyte phytochromes has a long N-terminal extension and a PCM followed by two PAS domains, a functional histidine kinase, and a REC domain (Fig. 5). This C-terminal architecture also is apparently ancestral to that of canonical plant phytochromes. The PAS repeat does not derive from the N-terminal PCM PAS domain, and the C-terminal histidine kinase is not descended from those of bacterial phytochromes (Duanmu et al., 2014). This architecture matches that of some PHYX1 proteins from streptophyte algae; while others lack the REC domain (Li et al., 2015). PHYX1 is the earliest-diverging lineage of streptophyte phytochromes (Fig. 3D), implicating the presence of phytochrome with this domain architecture in the common ancestor of prasinophytes and streptophytes. The REC domain is absent in the next streptophyte lineage to diverge, PHYX2 (Fig. 5). Interestingly, PHYX1 lacks a number of conserved residues required for chromophorylation and/or photoconversion (Li et al., 2015), so it may not function as a photoreceptor. However, PHYX1 and PHYX2 seem to be found together in algal transcriptomes. It thus seems plausible that these two proteins may function as an obligate heterodimer, which would explain the correlated deletion of both phytochromes. Similarly, it is known that Arabidopsis PHYC primarily functions as a heterodimer with PHYB (Clack et al., 2009).
Like streptophyte PHYX1, PHYX2 retains the phosphoacceptor His residue (Fig. 5). That residue was subsequently lost in the streptophyte PHY1/2 lineages, a large clade including phytochromes from early-diverging streptophyte algae and canonical plant phytochromes (Wickett et al., 2014; Li et al., 2015). The resulting C-terminus retains the PAS repeat of cryptophyte and prasinophyte phytochromes and contains both of the structural regions found in a functional histidine kinase, but canonical plant phytochromes lack the phosphoacceptor His residue (Fig. 5). Although such phytochromes exhibit Ser-Thr kinase activity (Yeh & Lagarias, 1998), the functional significance of this activity remains a point of ongoing debate (Ni et al., 2017; Pham et al., 2018). It is also possible that this controversy does not have a single answer. For example, some phytochromes may function to phosphorylate a subset of PIFs, but recruit other protein kinases in other cases.
Further phytochrome diversification occurred in streptophytes as they began to colonize land environments. For example, the moss Ceratodon purpureus contains an unusual phytochrome somewhat similar to a PEK, with a C-terminal eukaryotic protein kinase domain apparently replacing the histidine kinase-related output region (Thummler et al., 1992). As noted above, some algae and plants contain chimeric neochrome photoreceptors first discovered in ferns (Suetsugu et al., 2005). In neochromes, the phytochrome PCM is fused to the flavin-based blue light receptor phototropin, resulting in a photoreceptor which regulates chloroplast dynamics in response to red light via the Ser/Thr kinase domain of phototropins. The evolution of canonical plant phytochromes thus involves repeated duplication to give rise to PHYX1, PHYX2, and canonical plant phytochrome, diversification with loss of the REC domain followed by loss of the phosphoacceptor His residue, and deletion of PHYX1 and PHYX2 in the earliest-diverging streptophyte lineages. Overall, a key driving force for phytochrome diversification is the placement of the photosensory PCM into different protein contexts over the course of evolution.
VI. DIVERSIFICATION: UNTYING THE RED/FAR-RED KNOT
Canonical plant phytochromes use the knotted, three-domain PCM to photoconvert between red- and far-red sensing states. The first phytochromes characterized from algae and cyanobacteria exhibited very similar red/far-red photocycles (Kidd & Lagarias, 1990; Hughes et al., 1997; Yeh et al., 1997), as did phytochrome from a member of the sister clade to all other known streptophytes, the streptophyte Mesostigma viride (Wickett et al., 2014; Rockwell et al., 2017). Bacteriophytochromes exhibited similar photocycles (Davis et al., 1999; Giraud et al., 2002), but these proteins are red-shifted relative to plant phytochromes because their BV chromophore has a longer conjugated system (Fig. 4). Such photocycles are also seen in BV-utilizing fungal and diatom phytochromes (Blumenstein et al., 2005; Froehlich et al., 2005; Fortunato et al., 2016). Conventional thinking holds that such phytochromes are representative of the entire family, implying that phytochrome photosensing measures the red/far-red ratio and requires a knotted three-domain PCM.
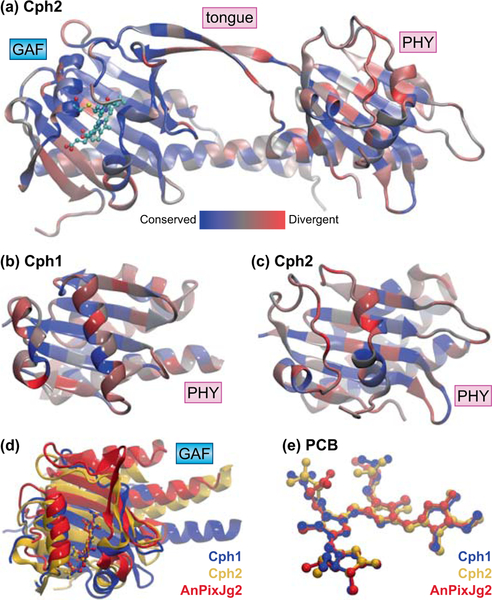
Modern research has overturned this view. Many cyanobacteria contain knotless phytochromes (also termed PAS-less or Cph2 phytochromes; Fig. 6). In such proteins, the N-terminal extension for the knot and the PAS domain are absent, yielding a two-domain GAF-PHY PCM (Wu & Lagarias, 2000; Anders et al., 2013). The large loop in the GAF domain is still present, even though there is no N-terminal extension for knot formation, and the red/far-red photocycle is quite similar to those of knotted phytochromes (Fig. 7). In the only available GAF-PHY structure for a knotless phytochrome, that for Cph2 from Synechocystis sp. PCC 6803 (Anders et al., 2013), the PHY domain adopts a slightly different fold relative to knotted cases of known structure (Fig. 6B–C). Interestingly, stramenopile phytochromes have a PHY domain apparently more closely related to those of knotless phytochromes than to those of other knotted phytochromes (Duanmu et al., 2014; Li et al., 2015), possibly indicating a similar structural adaptation. Deletion of this domain has produced varying results: some knotless phytochromes require the PHY domain for efficient formation of the photoproduct state, but others do not (Wu & Lagarias, 2000; Ulijasz et al., 2008; Gan et al., 2014).
Figure 6. Structures of knotless phytochromes and CBCRs.
(a) The GAF-PHY PCM of Cph2 from Synechocystis sp. PCC 6803 is shown (Anders et al., 2013), color-coded by sequence similarity using a published sequence alignment (Gan et al., 2014) as in Fig. 2(a). (b) Detailed view of the PHY domain of Cph1, colored as in (a); the tongue region and the long helices connecting to adjacent domains are omitted for clarity. (c) Detailed view of the PHY domain of Cph2, colored as in (a) and presented as in (b). (d) The bilin-binding GAF domain of AnPixJg2 (red) from Nostoc sp. PCC 7120 (sometimes designated Anabaena sp. PCC 7120) is compared to that of Cph2 (gold) from (a) and that of Cph1 (blue) from Fig. 2 (Anders et al., 2013; Narikawa et al., 2013), with PCB coordinates used for superposition. (e) The PCB chromophores and ligated Cys residues are shown superposed for Cph1 (red), Cph2 (blue), and AnPixJg2 (grey). All three cases correspond to dark-adapted, red-absorbing photostates in the C5–Z,syn C10–Z,syn C15–Z,anti configuration. Domain names: GAF, cGMP phosphodiesterase/Adenylate cyclase/FhlA; PHY, phytochrome-specific. PCB, phycocyanobilin; tongue, conserved insertion loop specific to the PHY domain.
Figure 7. Diversification of the phytochrome superfamily in cyanobacteria.
Jellybean domain architectures and photocycles are shown for selected photoreceptors (Wu & Lagarias, 2000; Rockwell et al., 2011; Rockwell et al., 2012a; Rockwell et al., 2012b; Savakis et al., 2012; Hirose et al., 2013; Gan et al., 2014; Rockwell et al., 2016). Photocycles are shown for heterologously expressed GAF-only constructs that are highlighted with blue shading in the domain architectures. In each case, the 15Z configuration is colored blue and the 15E configuration is colored orange; peak wavelengths and domain names are indicated. In the domain architectures, bilin-binding GAF domains are colored by photocycle, with the dark-adapted state on the left and the photoproduct on the right. Bilin chromophores are shown as blue polygons; domains that do not photoconvert (Rockwell et al., 2012b) are indicated with a white “photoproduct” state. The bilin-binding GAF lineage defining the phytochrome superfamily is only a subset of all GAF domains, so non-photosensory GAF domains are indicated in white. Domain names: GAF, cGMP phosphodiesterase/Adenylate cyclase/FhlA; PAS, Per/ARNT/Sim; PHY, phytochrome-specific; (H)kinase, histidine kinase bidomain, with the presence of the His indicated by H; GGDEF, diguanylate cyclase; HAMP, histidine kinase/adenylate cyclase/methyl-accepting chemotaxis protein/phosphatase; CBS, cystathionine β-synthase domain; EAL, diguanylate phosphodiesterase; MA-MCP, methyl-accepting domain of methylated chemotaxis proteins. Vertical rectangles on TePixJ are predicted transmembrane helices. DXCF, ins-C are different second Cys residues.
There are also knotted phytochromes that violate the red/far-red paradigm. Exceptions to this paradigm were first noted in phytochromes from the anoxygenic photosynthetic bacteria R. palustris and Bradyrhizobium sp. strain ORS278 (Giraud et al., 2005; Jaubert et al., 2007). Subsequently, N. punctiforme was shown to encode a very unusual phytochrome termed NpF1183 or TP1 (Rockwell et al., 2011). NpF1183 exhibits a violet-sensing dark state (peak absorption at 392 nm); photoconversion yielded an orange-absorbing species that underwent slow thermal conversion to a red-absorbing state (598 and 670 nm, respectively). This trichromatic behavior requires a second Cys residue immediately C-terminal to the GAF Cys. A similar tandem-Cys phytochrome from the glaucophyte C. paradoxa was shown to pair a blue-absorbing dark state with a conventional far-red sensing photoproduct (Rockwell et al., 2014). The phytochrome GwitGPS1, from the glaucophyte Gloeochaete wittrockiana, instead combines a conventional red-absorbing dark state with a novel blue-absorbing photoproduct state (Rockwell et al., 2014). Phytochromes from prasinophyte alga were also found to go beyond the expected red/far-red photocycle. All prasinophyte phytochromes characterized to date form a normal far-red absorbing 15E photoproduct, but this is associated with variable 15Z dark-adapted states absorbing yellow, orange, or red light ranging from 584 to 648 nm (Rockwell et al., 2014).
Perhaps the most surprising result to come out of these studies is the detection of blue or violet by phytochromes, because their chromophores intrinsically absorb red to green wavelengths of light (Lamparter & Michael, 2005; Rockwell et al., 2012a; Narikawa et al., 2015). The tandem Cys arrangement of NpF1183 was critical in answering this question. Replacement of the second Cys with Ala in NpF1183 and introduction of this residue into Cph1 demonstrated that the second Cys is necessary and sufficient for detection of short-wavelength light (Rockwell et al., 2011). Such tandem-Cys phytochromes are found in cyanobacterial genomes, glaucophyte transcriptomes, and transcriptomes from cryptophytes of the genus Hemiselmis (Keeling et al., 2014; Johnson et al., 2019). The tandem-Cys motif is absent in GwitGPS1, but this protein and related proteins from G. wittrockiana have a different conserved Cys residue proximal to the bilin-binding pocket (Rockwell et al., 2014). Thus, all known phytochromes detecting ultraviolet to blue wavelength light (370–470 nm) also have a second Cys residue. The function of such second Cys residues has been well established in CBCRs.
CBCRs have only been found in cyanobacteria and were discovered long after plant phytochromes (Butler et al., 1959; Kehoe & Grossman, 1996; Ikeuchi & Ishizuka, 2008; Fushimi & Narikawa, 2019). CBCRs have a bilin-binding GAF domain homologous to that of the phytochrome PCM (Fig. 6D–E) that includes the GAF Cys residue (Fig. 2). No other domains are required for folding, initial chromophore binding, covalent attachment, or photoconversion (Ikeuchi & Ishizuka, 2008). Indeed, one CBCR lineage consists of only an isolated bilin-binding GAF domain, yet it exhibits a conserved blue/orange photocycle (Rockwell et al., 2015b). Other CBCR domains can be found as one or more photosensory components of larger proteins, or even as long tandem arrays (Fig. 7). A number of CBCR subfamilies have been described to date via in vitro characterization and phylogenetic analysis (Hirose et al., 2008; Narikawa et al., 2008; Rockwell et al., 2011; Rockwell et al., 2012a; Rockwell et al., 2012b; Narikawa et al., 2014; Rockwell et al., 2015a; Rockwell et al., 2015b; Fushimi et al., 2016; Rockwell et al., 2016). These proteins exhibit an astonishing array of photocycles, with peak absorption for dark-adapted CBCRs ranging from circa 380–740 nm (ultraviolet to far-red) and that for their photoproducts ranging from circa 430–660 nm (blue to red). This diversity is achieved through several tuning mechanisms, including chromophore deprotonation, shortening of the bilin π system via isomerization at C5, and trapping of twisted intermediates (Ishizuka et al., 2011; Rockwell et al., 2012a; Hirose et al., 2013; Lim et al., 2018; Wiebeler et al., 2019).
Studies of CBCRs have also elucidated the basis for detection of violet or blue light. Every known CBCR photostate detecting light in the UV to blue (330–470 nm) requires a second Cys residue. In two such cases, it has been established that this residue forms a second thioether linkage to the C10 atom of the bilin chromophore in the blue- or violet-absorbing state (Ishizuka et al., 2011; Rockwell et al., 2011; Burgie et al., 2013; Lim et al., 2014). This second linkage shortens the conjugated system and blue-shifts the peak absorption. Given the presence of second Cys residues in all phytochromes known to sense this wavelength range, it seems likely that a similar tuning mechanism is at work in those proteins.
VII. DIVERSIFICATION: SPECIALIZED CIRCUITS AND MASTER REGULATORS
Our view of phytochrome has been shaped by its importance in development and shade avoidance for crop plants such as wheat and rice, annuals that grow rapidly from small seeds under the right conditions (Mathews, 2006; Casal, 2013). Plants that take different strategies, such as conifers, can behave differently during early development (Mathews, 2006; Mathews & Tremonte, 2012). Little is known about the function of canonical phytochromes in early-diverging streptophyte algae, much less about the function of PHYX1 and PHYX2. Phytochrome accumulates in the nucleus in response to light in the prasinophyte alga Micromonas pusilla as well as in flowering plants, so it seems likely that some aspects of phytochrome signaling are broadly conserved in Viridiplantae (Duanmu et al., 2014).
However, the biological function of phytochromes has clearly diversified from its ancestral state. Fungal phytochrome signaling proceeds not through PIFs but via the white collar complex and the HogA/Hog1 MAP kinase pathway (Yu et al., 2016). In cyanobacteria, a knotless phytochrome has been transferred to diverse cyanobacteria as part of a genomic island that permits growth in far-red light (Gan et al., 2014). This island induces a profound red shift in both light-harvesting and photosynthetic apparatus, with induction of ~900 genes and repression of ~2000 genes. Core subunits of photosystem I, photosystem II, and phycobilisomes are replaced by members of this 21-gene island. This far-red acclimation response depends on the function of the knotless phytochrome RfpA and its cognate response regulators (Zhao et al., 2015). R. palustris also uses phytochromes to regulate expression of light-harvesting pigments via dedicated signal transduction pathways involving histidine kinases and protein-protein interactions (Giraud et al., 2002; Giraud et al., 2005). In both cases, a small number of transcription factors are involved rather than the panoply of PIFs found in flowering plants (Pham et al., 2018).
We are also beginning to learn about the function of CBCRs (Wiltbank & Kehoe, 2019). The first known regulatory photobiological response for any cyanobacterium was complementary chromatic acclimation (CCA), described in the early 20th Century (Gaidukov, 1902). Over ninety years later, CCA was shown to be controlled by the green/red CBCRs RcaE and CcaS via dedicated histidine kinase pathways (Kehoe & Grossman, 1996; Hirose et al., 2008; Hirose et al., 2013). Other CBCRs are phototaxis sensors, as has been verified for SyPixJ1/TaxD in Synechocystis sp. PCC 6803 (Yoshihara et al., 2000; Bhaya et al., 2001), PixJSe in Synechococcus elongatus UTEX 3055 (Yang et al., 2018), and PtxD/NpF2164 in N. punctiforme (Campbell et al., 2015). These proteins contain one or more CBCR domains and a C-terminal methyl-accepting chemotaxis protein (MCP) domain. Others, like UirS/PixA, have C-terminal histidine kinases and modulate negative phototaxis via a phosphotransfer cascade (Narikawa et al., 2011; Song et al., 2011). In Thermosynechococcus elongatus and T. vulcanus, three CBCRs with GGDEF and/or EAL domains regulate cyclic-di-GMP levels in response to blue and green light, resulting in a switch between motile and sessile lifestyles (Enomoto et al., 2014; Enomoto et al., 2015). A CBCR from Microcoleus sp. PCC 7113 (cPAC or Mic7113_2205) has also been found to act as a light-regulated adenylate cyclase both in vitro and upon heterologous bacterial expression (Blain-Hartung et al., 2018).
We can thus see that the phytochrome superfamily encompasses far more than canonical plant phytochromes, with a complex, ongoing interplay between deletion, duplication, and diversification during evolution of life. Extant diversity has been shaped by a drive toward smaller knotless phytochromes and CBCRs in cyanobacteria, by spectral diversification from the ancestral red/far-red photoreceptor, by fusion to diverse domains in prokaryotes and eukaryotes, and by acquisition of different biological functions. Indeed, canonical plant phytochromes and PIFs seem to be the exception rather than the rule in phytochrome signaling. The greater diversity of algal phytochromes may also reflect the greater environmental and ecological diversity of eukaryotic algae compared to land plants.
VIII. AND NOW A WORD FROM THE LETTER E: ENVIRONMENT
The three themes of phytochrome evolution all interact with the environment of the organisms in which phytochromes are evolving, even though we may not understand the interactions. For example, deletion and diversification of phytochrome do not always correlate with photosynthesis. In one ochrophyte lineage, the absence of phytochrome roughly correlates with loss of photosynthesis (Beisser et al., 2017). By contrast, at least one non-photosynthetic Cryptomonas strain retains phytochromes (Keeling et al., 2014; Johnson et al., 2019). Photosynthetic organisms that have selected for small cell size and streamlined genomes sometimes lack phytochrome, as in the picoprasinophyte Ostreococcus but not in the related Micromonas pusilla (Palenik et al., 2007; Duanmu et al., 2014). Duplication and diversification of phytochromes and CBCRs seems roughly correlated with biological complexity in cyanobacteria. For example, there are only five CBCRs in T. elongatus, each of which has a single photosensory domain (Ikeuchi & Ishizuka, 2008), whereas N. punctiforme has 21 such proteins (Rockwell et al., 2015a). Many of these proteins have multiple photosensory domains in tandem, for a total of 41 phytochromes or CBCRs in the genome. N. punctiforme also exhibits much more complexity than T. elongatus: it exhibits CCA, nitrogen fixation, and differentiation of vegetative cells into three distinct, mutually exclusive cell types (Campbell et al., 2007; Hirose et al., 2010).
Diversification of phytochromes and CBCRs is likely driven by environmental factors. For example, consider competition between land plants and soil cyanobacteria. Land plants compete for sunlight via the shade avoidance response (Fig. 1). Given that the conifer Sequoia sempervirens can exceed 115 m in height, soil cyanobacteria cannot compete using a similar vertical response. However, CCA allows them to use green light that plants cannot, providing a successful alternative in shaded environments. CCA requires metabolically expensive remodeling of the light-harvesting apparatus (Kehoe & Gutu, 2006), so there is a clear selective advantage for regulating CCA with photosensors such as CcaS and RcaE. Photosynthetic organisms living in a water column face a different set of environmental challenges. Red and far-red light penetrate poorly in water, and there is thought to be some correlation between peak dark-state absorption of algal phytochromes and the depths at which those algae are encountered (Forest, 2014; Rockwell et al., 2014). Factors other than light can affect phytochrome function as well. The thermal instability of the photoproduct state confers a second sensory function upon phytochromes: dark reversion is a light-independent thermal reaction, so it proceeds with a pronounced temperature dependence in the presence or absence of light. The amount of Pfr that is present is thus dependent on both light and temperature, permitting phytochromes to exhibit different temperature responses depending on their dark reversion rates. Consistent with this, phytochromes are now known to function as temperature sensors in both prokaryotes and eukaryotes(Njimona & Lamparter, 2011; Njimona et al., 2014; Jung et al., 2016; Legris et al., 2016). Some CBCRs have even more unstable photoproducts and serve as broad-spectrum sensors of light intensity (Rockwell et al., 2012b). Although the evolutionary trajectory of this transition is not known, these observations demonstrate the extent to which the phytochrome superfamily provides photosensors for almost every color and purpose imaginable.
IX. CONCLUSION
Given the biological, photochemical, and ecological complexity of the phytochrome superfamily, phytochrome presents far more than a particularly vexatious exercise in phylogenetic inference. Indeed, each duplication and each fusion to a new C-terminus is a discrete evolutionary event with potential biological implications. Phytochromes are but one piece of a larger jigsaw puzzle for any organism: in some cases, they may be left in the box when the puzzle is complete, leading to their deletion, but in others they are critical. Deletion, duplication, and diversification have all played a part in shaping extant phytochromes as a whole and canonical plant phytochrome as a special case. In the future, we may expect new insights from improved sampling of biological diversity, from studies of phytochrome signaling mechanisms, from studies of chromophore biosynthesis, and from studies of bilin-based light harvesting systems. We also hope to learn more about the roles phytochromes play in the adaptation of photosynthetic organisms to changing environments brought on by transient breezes, daily and annual changes in solar irradiance, and the long-term effects of climate change. Sixty years after the first description of canonical plant phytochromes, the future of phytochrome science remains bright.
ACKNOWLEDGEMENTS
Work in the Lagarias lab supported by NIH grant R01 GM068552, by a grant from the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, United States Department of Energy (DOE DE-FG02–09ER16117) and by the USDA National Institute of Food and Agriculture, Hatch project number CA-D*-MCB-4126-H. We wish to thank Sarah Mathews for helpful discussions and for critical reading of the manuscript.
REFERENCES
- Anders K, Daminelli-Widany G, Mroginski MA, von Stetten D, Essen LO. 2013. Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. Journal of Biological Chemistry 288(50): 35714–35725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306(5693): 79–86. [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Forest KT. 2011. Bacterial phytochromes: more than meets the light. Critical Reviews in Biochemistry and Molecular Biology. 46(1): 67–88. [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Pierik R. 2017. The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell and Environment 40(11): 2530–2543. [DOI] [PubMed] [Google Scholar]
- Beisser D, Graupner N, Bock C, Wodniok S, Grossmann L, Vos M, Sures B, Rahmann S, Boenigk J. 2017. Comprehensive transcriptome analysis provides new insights into nutritional strategies and phylogenetic relationships of chrysophytes. PeerJ 5: e2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Takahashi A, Grossman AR. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proceedings of the National Academy of Sciences 98(13): 7540–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD. 2001. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414(6865): 776–779. [DOI] [PubMed] [Google Scholar]
- Blain-Hartung M, Rockwell NC, Moreno MV, Martin SS, Gan F, Bryant DA, Lagarias JC. 2018. Cyanobacteriochrome-based photoswitchable adenylyl cyclases (cPACs) for broad spectrum light regulation of cAMP levels in cells. Journal of Biological Chemistry 293(22): 8473–8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstein A, Vienken K, Tasler R, Purschwitz J, Veith D, Frankenberg-Dinkel N, Fischer R. 2005. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Current Biology 15(20): 1833–1838. [DOI] [PubMed] [Google Scholar]
- Brown MW, Heiss AA, Kamikawa R, Inagaki Y, Yabuki A, Tice AK, Shiratori T, Ishida KI, Hashimoto T, Simpson AGB, et al. 2018. Phylogenomics places orphan protistan lineages in a novel eukaryotic super-group. Genome Biology and Evolution 10(2): 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD. 2014a. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proceedings Of The National Academy Of Sciences 111(28): 10179–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Walker JM, Phillips GN Jr., Vierstra RD. 2013. A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 21(1): 88–97. [DOI] [PubMed] [Google Scholar]
- Burgie ES, Wang T, Bussell AN, Walker JM, Li H, Vierstra RD. 2014b. Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion. Journal of Biological Chemistry 289(35): 24573–24587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Zhang J, Vierstra RD. 2016. Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24(3): 448–457. [DOI] [PubMed] [Google Scholar]
- Burki F, Kaplan M, Tikhonenkov DV, Zlatogursky V, Minh BQ, Radaykina LV, Smirnov A, Mylnikov AP, Keeling PJ. 2016. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proceedings of the Royal Society B-Biological Sciences 283(1823). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler WL, Norris KH, Seigelman HW, Hendricks SB. 1959. Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proceedings Of The National Academy Of Sciences 45: 1703–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EL, Hagen KD, Chen R, Risser DD, Ferreira DP, Meeks JC. 2015. Genetic analysis reveals the identity of the photoreceptor for phototaxis in hormogonium filaments of Nostoc punctiforme. Journal of Bacteriology 197(4): 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EL, Summers ML, Christman H, Martin ME, Meeks JC. 2007. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. Journal of Bacteriology 189(14): 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. 2013. Photoreceptor signaling networks in plant responses to shade. Annual Review of Plant Biology 64: 403–427. [DOI] [PubMed] [Google Scholar]
- Chen A, Li C, Hu W, Lau MY, Lin H, Rockwell NC, Martin SS, Jernstedt JA, Lagarias JC, Dubcovsky J. 2014. PHYTOCHROME C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proceedings of the National Academy of Sciences 111(28): 10037–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J. 2011. Phytochrome signaling mechanisms and the control of plant development. Trends in Cell Biology 21(11): 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Shokry A, Moffet M, Liu P, Faul M, Sharrock RA. 2009. Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21(3): 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouze P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, et al. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465(7298): 617–621. [DOI] [PubMed] [Google Scholar]
- Collén J, Porcel B, Carre W, Ball SG, Chaparro C, Tonon T, Barbeyron T, Michel G, Noel B, Valentin K, et al. 2013. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proceedings of the National Academy of Sciences 110(13): 5247–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey HA, Dailey TA, Gerdes S, Jahn D, Jahn M, O’Brian MR, Warren MJ. 2017. Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiology and Molecular Biology Reviews 81(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Vener AV, Vierstra RD. 1999. Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 286(5449): 2517–2520. [DOI] [PubMed] [Google Scholar]
- Derelle R, Lopez-Garcia P, Timpano H, Moreira D. 2016. A phylogenomic framework to study the diversity and evolution of Stramenopiles (=Heterokonts). Molecular Biology and Evolution 33(11): 2890–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Bachy C, Sudek S, Wong CH, Jimenez V, Rockwell NC, Martin SS, Ngan CY, Reistetter EN, van Baren MJ, et al. 2014. Marine algae and land plants share conserved phytochrome signaling systems. Proceedings of the National Academy of Sciences 111: 15827–15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Casero D, Dent RM, Gallaher S, Yang W, Rockwell NC, Martin SS, Pellegrini M, Niyogi KK, Merchant SS, et al. 2013. Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proceedings of the National Academy of Sciences 110(9): 3621–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto G, Ni Ni W, Narikawa R, Ikeuchi M. 2015. Three cyanobacteriochromes work together to form a light color-sensitive input system for c-di-GMP signaling of cell aggregation. Proceedings Of The National Academy Of Sciences 112(26): 8082–8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto G, Nomura R, Shimada T, Win NN, Narikawa R, Ikeuchi M. 2014. Cyanobacteriochrome SesA is a diguanylate cyclase that induces cell aggregation in Thermosynechococcus. Journal of Biological Chemistry 289: 24801–24809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen LO, Mailliet J, Hughes J. 2008. The structure of a complete phytochrome sensory module in the Pr ground state. Proceedings of the National Academy of Sciences 105(38): 14709–14714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff NV, Battisti DS, Beachy RN, Cooper PJ, Fischhoff DA, Hodges CN, Knauf VC, Lobell D, Mazur BJ, Molden D, et al. 2010. Radically rethinking agriculture for the 21st century. Science 327(5967): 833–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Rockwell NC, Jang AY, Ernst LA, Waggoner AS, Duan Y, Lei H, Lagarias JC. 2005. Multiple roles of a conserved GAF domain tyrosine residue in cyanobacterial and plant phytochromes. Biochemistry 44(46): 15203–15215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixen KR, Baker AW, Stojkovic EA, Beatty JT, Harwood CS. 2014. Apo-bacteriophytochromes modulate bacterial photosynthesis in response to low light. Proceedings of the National Academy of Science 111(2): E237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest KT. 2014. Vivid watercolor paintbox for eukaryotic algae. Proceedings of the National Academy of Sciences 111(15): 5448–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato AE, Jaubert M, Enomoto G, Bouly JP, Raniello R, Thaler M, Malviya S, Bernardes JS, Rappaport F, Gentili B, et al. 2016. Diatom phytochromes reveal the existence of far-red-light-based sensing in the ocean. Plant Cell 28(3): 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg N, Mukougawa K, Kohchi T, Lagarias JC. 2001. Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell 13(4): 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. 2010. Phytochrome functions in Arabidopsis development. Journal of Experimental Botany 61(1): 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich AC, Noh B, Vierstra RD, Loros J, Dunlap JC. 2005. Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryotic Cell 4(12): 2140–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhang X, Yoshida T. 2004. Essential amino acid residues controlling the unique regioselectivity of heme oxygenase in Pseudomonas aeruginosa. Journal of the American Chemical Society 126(14): 4466–4467. [DOI] [PubMed] [Google Scholar]
- Fushimi K, Miyazaki T, Kuwasaki Y, Nakajima T, Yamamoto T, Suzuki K, Ueda Y, Miyake K, Takeda Y, Choi JH, et al. 2019. Rational conversion of chromophore selectivity of cyanobacteriochromes to accept mammalian intrinsic biliverdin. Proceedings of the National Academy of Sciences 116(17): 8301–8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K, Narikawa R. 2019. Cyanobacteriochromes: photoreceptors covering the entire UV-to-visible spectrum. Current Opinion in Structural Biology 57: 39–46. [DOI] [PubMed] [Google Scholar]
- Fushimi K, Rockwell NC, Enomoto G, Ni Ni W, Martin SS, Gan F, Bryant DA, Ikeuchi M, Lagarias JC, Narikawa R. 2016. Cyanobacteriochrome photoreceptors lacking the canonical Cys residue. Biochemistry 55(50): 6981–6995. [DOI] [PubMed] [Google Scholar]
- Gaidukov N. 1902. Über den Einfluss farbigen Lichts auf die Färbung lebender Oscillarien. Abhandlung Der Königlich Preussischen Akademie Der Wissenschaften 5: 1–36. [Google Scholar]
- Gan F, Zhang S, Rockwell NC, Martin SS, Lagarias JC, Bryant DA. 2014. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345(6202): 1312–1317. [DOI] [PubMed] [Google Scholar]
- Gawryluk RMR, Tikhonenkov DV, Hehenberger E, Husnik F, Mylnikov AP, Keeling PJ. 2019. Non-photosynthetic predators are sister to red algae. Nature 572(7768): 240–243. [DOI] [PubMed] [Google Scholar]
- Giraud E, Fardoux J, Fourier N, Hannibal L, Genty B, Bouyer P, Dreyfus B, Vermeglio A. 2002. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 417: 202–205. [DOI] [PubMed] [Google Scholar]
- Giraud E, Zappa S, Vuillet L, Adriano JM, Hannibal L, Fardoux J, Berthomieu C, Bouyer P, Pignol D, Vermeglio A. 2005. A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. Journal of Biological Chemistry 280(37): 32389–32397. [DOI] [PubMed] [Google Scholar]
- Gisk B, Wiethaus J, Aras M, Frankenberg-Dinkel N. 2012. Variable composition of heme oxygenases with different regiospecificities in Pseudomonas species. Archives of Microbiology 194(7): 597–606. [DOI] [PubMed] [Google Scholar]
- Glazer AN. 1985. Light harvesting by phycobilisomes. Annual Review of Biophysics and Biophysical Chemistry 14: 47–77. [DOI] [PubMed] [Google Scholar]
- Han YJ, Kim HS, Kim YM, Shin AY, Lee SS, Bhoo SH, Song PS, Kim JI. 2010a. Functional characterization of phytochrome autophosphorylation in plant light signaling. Plant Cell Physiology 51(4): 596–609. [DOI] [PubMed] [Google Scholar]
- Han YJ, Kim HS, Song PS, Kim JI. 2010b. Autophosphorylation desensitizes phytochrome signal transduction. Plant Signaling & Behavior 5(7): 868–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Narikawa R, Katayama M, Ikeuchi M. 2010. Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proceedings of the National Academy of Sciences 107(19): 8854–8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Rockwell NC, Nishiyama K, Narikawa R, Ukaji Y, Inomata K, Lagarias JC, Ikeuchi M. 2013. Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proceedings of the National Academy of Sciences 110(13): 4974–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Shimada T, Narikawa R, Katayama M, Ikeuchi M. 2008. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proceedings of the National Academy of Sciences 105(28): 9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC. 2013. Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proceedings of the National Academy of Sciences 110(4): 1542–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Su YS, Lagarias JC. 2009. A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Molecular Plant 2(1): 166–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. 2013. Phytochrome cytoplasmic signaling. Annual Review of Plant Biology 64: 377–402. [DOI] [PubMed] [Google Scholar]
- Hughes J, Lamparter T, Mittmann F, Hartmann E, Gärtner W, Wilde A, Borner T. 1997. A prokaryotic phytochrome. Nature 386(6626): 663. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Ishizuka T. 2008. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochemical & Photobiological Sciences 7(10): 1159–1167. [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Kamiya A, Suzuki H, Narikawa R, Noguchi T, Kohchi T, Inomata K, Ikeuchi M. 2011. The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry 50(6): 953–961. [DOI] [PubMed] [Google Scholar]
- Jaubert M, Lavergne J, Fardoux J, Hannibal L, Vuillet L, Adriano JM, Bouyer P, Pignol D, Giraud E, Vermeglio A. 2007. A singular bacteriophytochrome acquired by lateral gene transfer. Journal of Biological Chemistry 282(10): 7320–7328. [DOI] [PubMed] [Google Scholar]
- Johnson LK, Alexander H, Brown CT. 2019. Re-assembly, quality evaluation, and annotation of 678 microbial eukaryotic reference transcriptomes. Gigascience 8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. 2016. Phytochromes function as thermosensors in Arabidopsis. Science 354(6314): 886–889. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. 2014. The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biology 12(6): e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe DM, Grossman AR. 1996. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273(5280): 1409–1412. [DOI] [PubMed] [Google Scholar]
- Kehoe DM, Gutu A. 2006. Responding to color: the regulation of complementary chromatic adaptation. Annual Review of Plant Biology 57: 127–150. [DOI] [PubMed] [Google Scholar]
- Kidd DG, Lagarias JC. 1990. Phytochrome from the green alga Mesotaenium caldariorum - purification and preliminary characterization. Journal of Biological Chemistry 265(12): 7029–7035. [PubMed] [Google Scholar]
- Kohchi T, Mukougawa K, Frankenberg N, Masuda M, Yokota A, Lagarias JC. 2001. The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. The Plant Cell 13(2): 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooss S, Lamparter T. 2017. Cyanobacterial origin of plant phytochromes. Protoplasma 254(1): 603–607. [DOI] [PubMed] [Google Scholar]
- Kräutler B. 2014. Phyllobilins--the abundant bilin-type tetrapyrrolic catabolites of the green plant pigment chlorophyll. Chemical Society Reviews 43(17): 6227–6238. [DOI] [PubMed] [Google Scholar]
- Kyndt JA, Fitch JC, Meyer TE, Cusanovich MA. 2005. Thermochromatium tepidum photoactive yellow protein/bacteriophytochrome/diguanylate cyclase: characterization of the PYP domain. Biochemistry 44(12): 4755–4764. [DOI] [PubMed] [Google Scholar]
- Kyndt JA, Fitch JC, Meyer TE, Cusanovich MA. 2007. The photoactivated PYP domain of Rhodospirillum centenum Ppr accelerates the recovery of the bacteriophytochrome domain after white light illumination. Biochemistry 46(28): 8256–8262. [DOI] [PubMed] [Google Scholar]
- Kyndt JA, Fitch JC, Seibeck S, Borucki B, Heyn MP, Meyer TE, Cusanovich MA. 2010. Regulation of the Ppr histidine kinase by light-induced interactions between its photoactive yellow protein and bacteriophytochrome domains. Biochemistry 49(8): 1744–1754. [DOI] [PubMed] [Google Scholar]
- Lagarias JC, Rapoport H. 1980. Chromopeptides from phytochrome. The structure and linkage of the Pr form of the phytochrome chromophore. Journal of the American Chemical Society 102: 4821–4828. [Google Scholar]
- LaMattina JW, Nix DB, Lanzilotta WN. 2016. Radical new paradigm for heme degradation in Escherichia coli O157:H7. Proceedings of the National Academy of Science 113(43): 12138–12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamparter T, Michael N. 2005. Agrobacterium phytochrome as an enzyme for the production of ZZE bilins. Biochemistry 44(23): 8461–8469. [DOI] [PubMed] [Google Scholar]
- Lax G, Eglit Y, Eme L, Bertrand EM, Roger AJ, Simpson AGB. 2018. Hemimastigophora is a novel supra-kingdom-level lineage of eukaryotes. Nature 564(7736): 410–414. [DOI] [PubMed] [Google Scholar]
- Lee DW, Graham R. 1986. Leaf optical properties of rainforest sun and extreme shade plants. American Journal of Botany 73(8): 1100–1108. [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A, Wigge PA, Schafer E, Vierstra RD, Casal JJ. 2016. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354(6314): 897–900. [DOI] [PubMed] [Google Scholar]
- Li FW, Melkonian M, Rothfels CJ, Villarreal JC, Stevenson DW, Graham SW, Wong GK, Pryer KM, Mathews S. 2015. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nature Communications 6: 7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FW, Villarreal JC, Kelly S, Rothfels CJ, Melkonian M, Frangedakis E, Ruhsam M, Sigel EM, Der JP, Pittermann J, et al. 2014. Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proceedings of the National Academy of Sciences 111(18): 6672–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Rockwell NC, Martin SS, Dallas JL, Lagarias JC, Ames JB. 2014. Photoconversion changes bilin chromophore conjugation and protein secondary structure in the violet/orange cyanobacteriochrome NpF2163g3. Photochemical & Photobiological Sciences 13(6): 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Yu Q, Gottlieb SM, Chang CW, Rockwell NC, Martin SS, Madsen D, Lagarias JC, Larsen DS, Ames JB. 2018. Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4. Proceedings of the National Academy of Science 115(17): 4387–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommer M, Specht M, Roy AS, Kraemer L, Andreson R, Gutowska MA, Wolf J, Bergner SV, Schilhabel MB, Klostermeier UC, et al. 2012. Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biology 13(7): R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S. 2006. Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Molecular Ecology 15(12): 3483–3503. [DOI] [PubMed] [Google Scholar]
- Mathews S, McBreen K. 2008. Phylogenetic relationships of B-related phytochromes in the Brassicaceae: redundancy and the persistence of phytochrome D. Molecular Phylogenetics and Evolution 49(2): 411–423. [DOI] [PubMed] [Google Scholar]
- Mathews S, Tremonte D. 2012. Tests of the link between functional innovation and positive selection at phytochrome A: the phylogenetic distribution of far-red high-irradiance responses in seedling development. International Journal of Plant Science 173(6): 662–672. [Google Scholar]
- Matsui T, Nambu S, Ono Y, Goulding CW, Tsumoto K, Ikeda-Saito M. 2013. Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide. Biochemistry 52(18): 3025–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JT, Lagarias JC. 1997. The phytofluors: a new class of fluorescent protein probes. Current Biology 7(11): 870–876. [DOI] [PubMed] [Google Scholar]
- Nagatani A. 2004. Light-regulated nuclear localization of phytochromes. Current Opinion in Plant Biology 7(6): 708–711. [DOI] [PubMed] [Google Scholar]
- Nambu S, Matsui T, Goulding CW, Takahashi S, Ikeda-Saito M. 2013. A new way to degrade heme: the Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO. Journal of Biological Chemistry 288(14): 10101–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narikawa R, Enomoto G, Win N-N, Fushimi K, Ikeuchi M. 2014. A new type of dual-cys cyanobacteriochrome GAF domain found in cyanobacterium Acaryochloris marina, which has an unusual red/blue reversible photoconversion cycle. Biochemistry 53(31): 5051–5059. [DOI] [PubMed] [Google Scholar]
- Narikawa R, Fukushima Y, Ishizuka T, Itoh S, Ikeuchi M. 2008. A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. Journal of Molecular Biology 380(5): 844–855. [DOI] [PubMed] [Google Scholar]
- Narikawa R, Ishizuka T, Muraki N, Shiba T, Kurisu G, Ikeuchi M. 2013. Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proceedings of the National Academy of Sciences 110(3): 918–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narikawa R, Nakajima T, Aono Y, Fushimi K, Enomoto G, Ni Ni W, Itoh S, Sato M, Ikeuchi M. 2015. A biliverdin-binding cyanobacteriochrome from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Scientific Reports 5: 7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narikawa R, Suzuki F, Yoshihara S, Higashi SI, Watanabe M, Ikeuchi M. 2011. Novel photosensory two-component system (PixA-NixB-NixC) involved in the regulation of positive and negative phototaxis of cyanobacterium synechocystis sp. PCC 6803. Plant and Cell Physiology 52(12): 2214–2224. [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, Gonzalez-Grandio E, Chalkley RJ, Huhmer AFR, Burlingame AL, Wang ZY, Quail PH. 2017. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nature Communications 8: 15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njimona I, Lamparter T. 2011. Temperature effects on Agrobacterium phytochrome Agp1. PLoS One 6(10): e25977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njimona I, Yang R, Lamparter T. 2014. Temperature effects on bacterial phytochrome. PLoS One 9(10): e109794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Yang S, Choi G. 2012. Phytochrome regulates translation of mRNA in the cytosol. Proceedings of the National Academy of Science 109(4): 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouze P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, et al. 2007. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proceedings of the National Academy of Sciences 104(18): 7705–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, Huq E. 2018. Phytochromes and phytochrome interacting factors. Plant Physiology 176(2): 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DC, Goodenough UW, Roth R, Lee JH, Kariyawasam T, Mutwil M, Ferrari C, Facchinelli F, Ball SG, Cenci U, et al. 2019. Analysis of an improved Cyanophora paradoxa genome assembly. DNA Research 26(4): 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Lee JM, Yoon HS, Bhattacharya D. 2017. Hypothesis: gene–rich plastid genomes in red algae may be an outcome of nuclear genome reduction. Journal of Phycology 53(3): 715–719. [DOI] [PubMed] [Google Scholar]
- Quest B, Hubschmann T, Sharda S, Tandeau de Marsac N, Gartner W. 2007. Homologous expression of a bacterial phytochrome. The cyanobacterium Fremyella diplosiphon incorporates biliverdin as a genuine, functional chromophore. FEBS J 274(8): 2088–2098. [DOI] [PubMed] [Google Scholar]
- Ratliff M, Zhu W, Deshmukh R, Wilks A, Stojiljkovic I. 2001. Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. Journal of Bacteriology 183(21): 6394–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA. 1969. Action spectra for photosynthesis and light-stimulated ion transport processes in Hydrodictyon africanum. New Phytologist 68: 45–62. [Google Scholar]
- Rockwell NC, Duanmu D, Martin SS, Bachy C, Price DC, Bhattacharya D, Worden AZ, Lagarias JC. 2014. Eukaryotic algal phytochromes span the visible spectrum. Proceedings of the National Academy of Sciences 111(10): 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC. 2006. The structure of phytochrome: a picture is worth a thousand spectra. Plant Cell 18(1): 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC. 2007. Flexible mapping of homology onto structure with homolmapper. BMC Bioinformatics 8: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC. 2017. Ferredoxin-dependent bilin reductases in eukaryotic algae: ubiquity and diversity. Journal of Plant Physiology 217: 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Feoktistova K, Lagarias JC. 2011. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proceedings of the National Academy of Sciences 108(29): 11854–11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Gan F, Bryant DA, Lagarias JC. 2015a. NpR3784 is the prototype for a distinctive group of red/green cyanobacteriochromes using alternative Phe residues for photoproduct tuning. Photochemical & Photobiological Sciences 14(2): 258–269. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. 2012a. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry 51(7): 1449–1463. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC. 2012b. Red/green cyanobacteriochromes: sensors of color and power. Biochemistry 51(48): 9667–9677. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC. 2015b. Identification of DXCF cyanobacteriochrome lineages with predictable photocycles. Photochemical & Photobiological Sciences 14(5): 929–941. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC. 2016. Identification of cyanobacteriochromes detecting far-red light. Biochemistry 55(28): 3907–3919. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Li FW, Mathews S, Lagarias JC. 2017. The phycocyanobilin chromophore of streptophyte algal phytochromes is synthesized by HY2. New Phytologist 214(3): 1145–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. 2006. Phytochrome structure and signaling mechanisms. Annual Review of Plant Biology 57: 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoni AW, Price DC, Seger M, Lyska D, Lammers P, Bhattacharya D, Weber APM. 2019. The genomes of polyextremophilic Cyanidiales contain 1% horizontally transferred genes with diverse adaptive functions. Elife 8. pii: e45017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Gomelsky M. 2014. Near-infrared light responsive synthetic c-di-GMP module for optogenetic applications. ACS Synthetic Biology 3(11): 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salewski J, Escobar FV, Kaminski S, von Stetten D, Keidel A, Rippers Y, Michael N, Scheerer P, Piwowarski P, Bartl F, et al. 2013. Structure of the biliverdin cofactor in the Pfr state of bathy and prototypical phytochromes. Journal of Biological Chemistry 288(23): 16800–16814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savakis P, De Causmaecker S, Angerer V, Ruppert U, Anders K, Essen LO, Wilde A. 2012. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Molecular Microbiology 85(2): 239–251. [DOI] [PubMed] [Google Scholar]
- Schroeder DC, Park Y, Yoon HM, Lee YS, Kang SW, Meints RH, Ivey RG, Choi TJ. 2009. Genomic analysis of the smallest giant virus--Feldmannia sp. virus 158. Virology 384(1): 223–232. [DOI] [PubMed] [Google Scholar]
- Song JY, Cho HS, Cho JI, Jeon JS, Lagarias JC, Park YI. 2011. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proceedings of the National Academy of Sciences 108(26): 10780–10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B, Sanchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdan PD. 2010. Arabidopsis thaliana life without phytochromes. Proceedings of the National Academy of Sciences 107(10): 4776–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassert JFH, Jamy M, Mylnikov AP, Tikhonenkov DV, Burki F. 2019. New phylogenomic analysis of the enigmatic phylum Telonemia further resolves the eukaryote tree of life. Molecular Biology and Evolution 36(4): 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N, Mittmann F, Wagner G, Hughes J, Wada M. 2005. A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proceedings Of The National Academy Of Sciences 102(38): 13705–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala H, Bjorling A, Berntsson O, Lehtivuori H, Niebling S, Hoernke M, Kosheleva I, Henning R, Menzel A, Ihalainen JA, et al. 2014. Signal amplification and transduction in phytochrome photosensors. Nature 509(7499): 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummler F, Dufner M, Kreisl P, Dittrich P. 1992. Molecular cloning of a novel phytochrome gene of the moss Ceratodon purpureus which encodes a putative light-regulated protein kinase. Plant Molecular Biology 20(6): 1003–1017. [DOI] [PubMed] [Google Scholar]
- Uenaka H, Kadota A. 2007. Functional analyses of the Physcomitrella patens phytochromes in regulating chloroplast avoidance movement. Plant Journal 51(6): 1050–1061. [DOI] [PubMed] [Google Scholar]
- Ulijasz AT, Cornilescu G, von Stetten D, Kaminski S, Mroginski MA, Zhang J, Bhaya D, Hildebrandt P, Vierstra RD. 2008. Characterization of two thermostable cyanobacterial phytochromes reveals global movements in the chromophore-binding domain during photoconversion. Journal of Biological Chemistry 283(30): 21251–21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez Escobar F, Buhrke D, Fernandez Lopez M, Shenkutie SM, von Horsten S, Essen LO, Hughes J, Hildebrandt P. 2017. Structural communication between the chromophore-binding pocket and the N-terminal extension in plant phytochrome phyB. FEBS Letters 591(9): 1258–1265. [DOI] [PubMed] [Google Scholar]
- Vuillet L, Kojadinovic M, Zappa S, Jaubert M, Adriano JM, Fardoux J, Hannibal L, Pignol D, Vermeglio A, Giraud E. 2007. Evolution of a bacteriophytochrome from light to redox sensor. EMBO Journal 26(14): 3322–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. 2005. A light-sensing knot revealed by the structure of the chromophore binding domain of phytochrome. Nature 438(7066): 325–331. [DOI] [PubMed] [Google Scholar]
- Wagner JR, Zhang J, von Stetten D, Gunther M, Murgida DH, Mroginski MA, Walker JM, Forest KT, Hildebrandt P, Vierstra RD. 2008. Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. Journal of Biological Chemistry 283(18): 12212–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proceedings of the National Academy of Sciences 111(45): E4859–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebeler C, Rao AG, Gärtner W, Schapiro I. 2019. The effective conjugation length is responsible for the red/green spectral tuning in the cyanobacteriochrome Slr1393g3. Angewandte Chemie-International Edition 58(7): 1934–1938. [DOI] [PubMed] [Google Scholar]
- Wiltbank LB, Kehoe DM. 2019. Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nature Reviews Microbiology 17(1): 37–50. [DOI] [PubMed] [Google Scholar]
- Wu SH, Lagarias JC. 2000. Defining the bilin lyase domain: lessons from the extended phytochrome superfamily. Biochemistry 39(44): 13487–13495. [DOI] [PubMed] [Google Scholar]
- Wu SH, McDowell MT, Lagarias JC. 1997. Phycocyanobilin is the natural precursor of the phytochrome chromophore in the green alga Mesotaenium caldariorum. Journal of Biological Chemistry 272(41): 25700–25705. [DOI] [PubMed] [Google Scholar]
- Yang X, Kuk J, Moffat K. 2008. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proceedings of the National Academy of Sciences 105(38): 14715–14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Lam V, Adomako M, Simkovsky R, Jakob A, Rockwell NC, Cohen SE, Taton A, Wang J, Lagarias JC, et al. 2018. Phototaxis in a wild isolate of the cyanobacterium Synechococcus elongatus. Proceedings of the National Academy of Science. 115(52): E12378–E12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Lagarias JC. 1998. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proceedings Of The National Academy Of Sciences 95: 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Wu S-H, Murphy JT, Lagarias JC. 1997. A cyanobacterial phytochrome two-component light sensory system. Science 277: 1505–1508. [DOI] [PubMed] [Google Scholar]
- Yoshihara S, Suzuki F, Fujita H, Geng XX, Ikeuchi M. 2000. Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiology 41(12): 1299–1304. [DOI] [PubMed] [Google Scholar]
- Yu Z, Armant O, Fischer R. 2016. Fungi use the SakA (HogA) pathway for phytochrome-dependent light signalling. Nature Microbiology 1: 16019. [DOI] [PubMed] [Google Scholar]
- Zhao C, Gan F, Shen G, Bryant DA. 2015. RfpA, RfpB, and RfpC are the master control elements of far-red light photoacclimation (FaRLiP). Frontiers in Microbiology 6: 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]