Figure 5. Diverse domain architectures in knotted phytochromes.
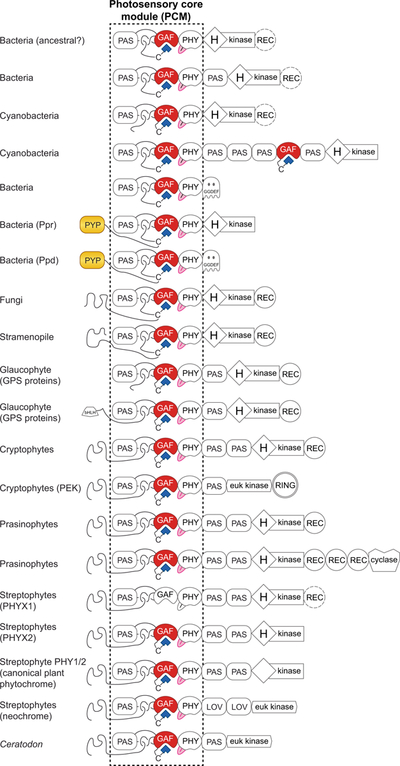
Jellybean domain diagrams are shown for diverse prokaryotic and eukaryotic phytochromes that use the canonical knotted PCM (Duanmu et al., 2014; Li et al., 2015). Bilin-binding GAF domains are red, the tongue region of the PHY domain is pink, bilin chromophores are shown as blue polygons, and chromophore-binding PYP domains are yellow. REC domains that are present in only some cases are dashed, as in PHYX1. Domain names: GAF, cGMP phosphodiesterase/Adenylate cyclase/FhlA; PAS, Per/ARNT/Sim; PHY, phytochrome-specific; (H)kinase, histidine kinase bidomain, with the presence of the His indicated by H; REC, response regulator receiver; RING, really interesting new gene; euk kinase, eukaryotic protein kinase; cyclase, eukaryotic adenylate/guanylate cyclase; GGDEF, diguanylate cyclase; PYP, photoactive yellow protein; bHLH, basic helix-loop-helix; LOV, PAS domain belonging to the light/oxygen/voltage lineage. Two-Cys phytochromes from cyanobacteria and glaucophytes are not indicated.
