Neuroradiology and imaging-based diagnostics in general have a dire need for scan techniques with improved microstructural sensitivity for detection of subtle tissue alterations in early disease phases or for diagnostics of diseases that are currently simply radiologically invisible. Diseases in which this need is most outspoken are often neurodegenerative diseases such as Alzheimer's disease, multiple sclerosis, and Parkinson's disease but examples also include (mild, repetitive) trauma, addiction, and stroke. Less frequently reported but important nevertheless is the need for sensitive methods for diagnosis and monitoring of patients exposed to poisonous substances. In the industrialized parts of the world, the leading cause of poisoning is carbon monoxide (CO).1 In the study by Zhang et al2 in the present issue of the American Journal of Neuroradiology (AJNR), brain microstructure in CO-poisoned patients is assessed at 3 time points using diffusional kurtosis imaging (DKI) and is correlated to patient cognitive performance. It concluded that DKI metrics provide important information about the damage to the brain due to CO, and supplement cognitive scores. As a first, the study found kurtosis fractional anisotropy (KFA) to have the best diagnostic efficiency based on a standard area under the curve measure. Because this parameter is likely to be somewhat new to many working in the field of neuroradiology, this commentary aims to recapitulate the basis of KFA and what it might indicate about tissue microstructure.
Diffusion MR Imaging and Diffusional Kurtosis Imaging
Water is abundant in the brain, and the water molecules are thermally driven to move ceaselessly and randomly. The local environment of a water molecule determines its mobility. Therefore, water ensemble properties such as mean diffusivity, diffusion anisotropy, and diffusion distribution shape vary among tissues with different compositions. In neural tissue, diffusion variations are seen among tissue microdomains at the subcellular level,3 the intra- or extracellular space, among cell types,4 and therefore also on a coarser scale between gray and white matter tissue.5 Noninvasive MR imaging–based measurements of brain-water diffusion, therefore, in principle, contain abundant information about the cellular-level tissue composition. Initially, the diffusion MR imaging (dMRI) signal description was based on a Gaussian (normal) diffusion probability distribution,6 forming the basis for the familiar diffusion tensor imaging technique. In biologic tissue, however, the overall diffusion behavior observed from an MR imaging voxel is not Gaussian under clinically relevant measurement conditions. To account for non-Gaussian diffusion, a kurtosis term is added to the DTI signal equation, producing the diffusion DKI framework:7
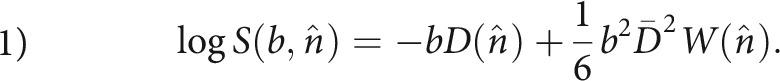 |
Equation 1 describes the behavior of the (log of the) normalized dMRI signal from tissue with diffusion described by a 3 × 3 diffusion tensor D, and kurtosis is described by the 4D tensor W. As written here, the signal is measured along a direction n̂ with diffusion weighting b. The first term on the right-hand side is the DTI signal term, so DTI is fully contained (and actually improved8) in DKI. The second term on the right-hand side of Equation 1 is the kurtosis term, with the kurtosis tensor W describing the non-Gaussian properties of the diffusion, which are not contained in D. As written here, D(n̂)is the apparent diffusivity and W(n̂) is the apparent kurtosis, both observed along the diffusion gradient direction n̂. Note that sometimes the apparent kurtosis is referred to as K(n̂), where K(n̂) = W(n̂)D̄2/D(n̂)2 (D̄ is mean diffusivity, see below). Simply put, the kurtosis term accounts for the signal deviation from log-linear DTI behavior along the diffusion-encoding direction n̂.
The wealth of information available from DTI and DKI is contained in the diffusion tensor D and the kurtosis tensor W. From D, a number of parameters are available, with typical reported metrics being the apparent diffusion coefficient, ADC = D(n̂); mean diffusivity (D̄ ≡ Tr(D)/3 = (λ1 + λ2 + λ3)/3, where λ1−3 is the diffusion tensor eigenvalues—that is, D(x̂′), D(ŷ′), D(ẑ′) in the tensor eigenframe); radial and axial diffusivities; and fractional anisotropy (FA).9 In white matter, the direction with the highest diffusivity (axial diffusivity) largely identifies the main fiber direction.10–12 The pronounced anisotropy of brain white matter is due to the myelin sheath, the axonal membrane itself, and the cytoskeleton inside the axon,13 which collectively cause WM FA to be high (typically >0.6, however see the example below). In DKI, D and W are used together to provide more parameters. These tensors will behave very differently: For example, in a single fiber system, the observed diffusion is high along the fiber direction and low across the fiber direction. In contrast, the kurtosis would be higher across than along the fiber direction, mostly due to restriction effects. This added information improves the capability of dMRI to detect changes in brain and body organ microstructure.14,15 From DKI, typically reported metrics are analogously the mean kurtosis (MK) and radial and axial kurtosis, and, for very aligned WM, a set of biomarkers from the WM tract integrity framework.16 More recently, the KFA has been introduced.17,18 These parameters, their estimation, and varying definitions are reviewed in Hansen and Jespersen.19
A comprehensive review of DKI in neuroimaging is outside the scope of this commentary, but to illustrate the potential of DKI, we list key neurologic disorders and neuroradiologic areas where DKI has already proved useful: addiction,20 stroke,21–23 Alzheimer's disease,24 multiple sclerosis,25 Parkinson's disease,26–29 brain cancer (gliomas),30,31 and head trauma/concussion.32–36 While DKI is increasingly used, so far the relatively new KFA has not been given much attention in preclinical and clinical studies. It is, therefore, noteworthy that KFA features prominently in the article of Zhang et al.2 Thus, KFA and its interpretation are the main focus of the remainder of this commentary.
dMRI Anisotropy Measures: FA and KFA
The DKI framework produces 2 scalar anisotropy measures: the FA from the diffusion tensor D and the KFA from the kurtosis tensor W. The FA is defined as
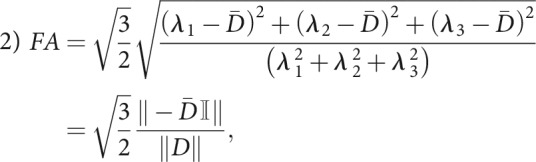 |
where double bars denote the Euclidean tensor norm (the Frobenius norm), 𝕀 is the 3 × 3 identity matrix, and the remaining parameters are defined above. The numeric front factor ensures that that the FA assumes values in the range from 0 (completely isotropic diffusion) to 1 (fully anisotropic, unidirectional diffusion). Although a few alternative definitions of the kurtosis anisotropy have been proposed (notably the one in Poot et al37), the agreed upon definition today is the one introduced in Hansen et al:18
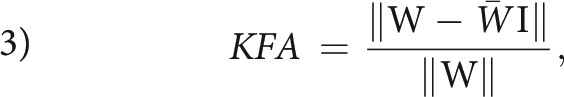 |
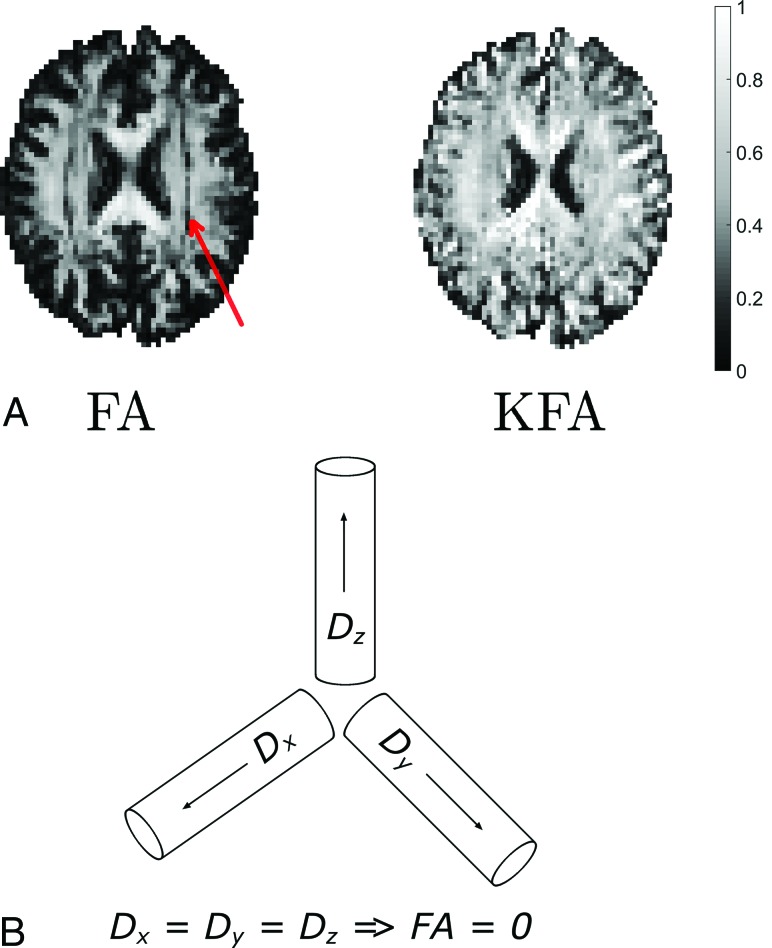
where W is the kurtosis tensor, W̄ is the kurtosis tensor mean,18,38,39 and I is the fully symmetric rank 4 isotropic tensor. Conveniently, this definition of KFA is analogous to the FA definition in Equation 2, with the only difference being due to tensor dimensions. We note the absence of a numeric front factor in Equation 3; KFA naturally assumes values in the 0–1 range. Interested readers are referred to early explorations of KFA in Hansen and Jespersen17 and Glenn et al.40 In these studies, simulations and experiments were used to investigate the information contained in the KFA. It was found that KFA contrast supplements the FA in important ways. This feature is illustrated in the Figure showing side-by-side maps of FA and KFA in the same image plane position in normal human brain (Fig 1A).
FIGURE.
A, Maps of FA and KFA in the same section position in the normal human brain. The color bar on the right applies to both parameter maps. In the FA map in A, a band of low FA values is seen as a dark band in both hemispheres. The red arrow points to this feature. The same white matter region is seen to have KFA values similar to those of the surrounding white matter. B, The fiber arrangement causing this FA behavior is illustrated in a simplified thought experiment. Figure adapted with permission from Hansen and Jespersen.17
The red arrow points to a dark band in the FA map inside the WM. This band is seen in both hemispheres (note the symmetry). In each hemisphere, this band comprises voxels with low FA values, though the voxels are located in anatomic WM. Conversely, in the KFA map on the right in Fig 1A, we see that KFA in WM is high and notably retains its high value in the regions where FA fails as a reporter of anisotropy. These WM voxels assume low FA values because they contain a crossing-fiber arrangement in which WM fibers along 3 orthogonal directions weave between each other. This effectively produces the situation illustrated in the schematic in Fig 1B, which shows 3 separate WM fiber bundles that intersect at right angles. In this example, we ignore extra-axonal diffusion. We see that if diffusivities in the 3 fibers are similar (as they are likely to be), the mean diffusivity will be the same as the eigenvalues (ie, the diffusivities along 3 fiber directions D̄ = (λ1 + λ2 + λ3)/3 ≈ λ1−3) causing the FA to vanish because, in this case, each term in the numerator in the middle expression in Equation 2 will be approximately zero. This happens despite the diffusivity in this fiber arrangement being highly anisotropic. Although not particularly complex, the anisotropy in this fiber arrangement cannot be described by the diffusion tensor, resulting in the low FA constituting the dark band in this WM region.
In the interpretation of these metrics, it is crucial to remember that FA and KFA report on different features of the diffusion process. We, therefore, stress that FA summarizes the spatial variation of diffusion rates, whereas KFA summarizes the directional variation in the degree of non-Gaussian diffusion. Loosely speaking, another difference is that the KFA stems from the kurtosis tensor, which is a 4D tensor with much more “room” to capture, in detail, the spatial variation in kurtosis. Collectively, these factors contribute to the demonstrated behavior in which KFA continues to provide contrast in areas where FA does not. Both metrics can be difficult to interpret in strict terms of tissue properties, but on the basis of the simple example above, we can cautiously state that KFA reflects tissue-diffusion complexity. In the example above, we saw that the 2D diffusion tensor cannot resolve anisotropy in voxels with complex fiber composition. However, we also saw that areas with complex fiber arrangements may be distinguished from genuine low-anisotropy regions using FA and KFA in combination: If FA is low and KFA is high, then likely the diffusion is, in fact, anisotropic but the fiber arrangement is too complex to be captured by the diffusion tensor. Thus, the KFA is a good parameter to include in studies in which subtle remodeling is expected in complex tissue regions. While KFA may therefore prove to be a valuable marker of tissue microstructure, we nevertheless stress that no DKI parameter is specifically bound to a particular microscopic tissue component.
Discussion and Future Perspectives
DKI is sometimes omitted from clinical protocols due to longer acquisition and parameter estimation times than a simple DTI protocol. While strategies exist for fast estimation of most DKI parameters,18,19,38–44 robust estimation of KFA still requires the full kurtosis tensor W to be determined by fitting on a pixel-by-pixel basis.17 This requires multishell dMRI data with typically 30 directions and 2–3 nonzero b-values. With modern dMRI techniques, this is possible in clinically feasible scan times.45 From such datasets, DKI parameter estimation is possible using any one of the many software packages that exist for dMRI data analysis such as MUSC's DKE software package (https://medicine.musc.edu/departments/centers/cbi/dki/dki-data-processing), which also computes the KFA as defined in Equation 3. As noted above, interpretation of DKI findings in terms of biophysical tissue properties is a difficult problem. In preclinical work, subsequent histology may be used to interpret DKI findings,15,46,47 and insights gained from such efforts may aid in interpretation of clinical DKI.
In the discussion of the example in the Figure, we noted that KFA somehow reflects the diffusion anisotropy below the voxel level. Other techniques achieve similar sensitivity48,49 but require nonstandard pulse sequences. Until such techniques reach the clinic in earnest, the KFA may serve as an indicator of the diagnostic potential of such sensitivity.
CONCLUSIONS
Imaging techniques with improved sensitivity to microstructure have utility in many areas of neuroimaging, including diagnosis and monitoring of patients exposed to toxic or poisonous substances. The study by Zhang et al2 uses the microstructurally sensitive DKI framework to assess cerebral damage in CO-poisoned subjects. From DKI, the authors obtain measures of mean diffusivity, FA, MK, and the less explored KFA. These measures are then correlated to neuropsychiatric scores. The work presents a timely contribution to 2 avenues of neuroradiology: 1) the exploration of DKI and KFA in clinical practice, and 2) identification of sensitive markers for diagnostics of CO-intoxicated patients. Insights gained from this study may also benefit and inspire many other areas of neuroimaging where the same techniques could be used. With this commentary, we hope that the KFA will have become more familiar to readers of AJNR so that investigators will consider it a parameter of interest in their future work.
References
- 1. Omaye ST. Metabolic modulation of carbon monoxide toxicity. Toxicology 2002;180:139–50 10.1016/S0300-483X(02)00387-6 [DOI] [PubMed] [Google Scholar]
- 2. Zhang Y, Wang T, Lei J, et al. . Cerebral damage after carbon monoxide poisoning: a longitudinal diffusional kurtosis imaging study. AJNR Am J Neuroradiol 2019;40:1630–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee CH, Flint JJ, Hansen B, et al. . Investigation of the subcellular architecture of L7 neurons of Aplysia Californica using magnetic resonance microscopy (MRM) at 7.8 microns. Sci Rep 2015;5:11147 10.1038/srep11147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flint JJ, Hansen B, Portnoy S, et al. . Magnetic resonance microscopy of human and porcine neurons and cellular processes. Neuroimage 2012;60:1404–11 10.1016/j.neuroimage.2012.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Flint JJ, Lee CH, Hansen B, et al. . Magnetic resonance microscopy of mammalian neurons. Neuroimage 2009;46:1037–40 10.1016/j.neuroimage.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin-echo. J Magn Reson B 1994;103:247–54 10.1006/jmrb.1994.1037 [DOI] [PubMed] [Google Scholar]
- 7. Jensen JH, Helpern JA, Ramani A, et al. . Diffusional kurtosis imaging: the quantification of non-Gaussian water diffusion by means of magnetic resonance imaging. Magn Reson Med 2005;53:1432–40 10.1002/mrm.20508 [DOI] [PubMed] [Google Scholar]
- 8. Veraart J, Poot DHJ, Van Hecke W, et al. . More accurate estimation of diffusion tensor parameters using diffusion kurtosis imaging. Magn Reson Med 2011;65:138–45 10.1002/mrm.22603 [DOI] [PubMed] [Google Scholar]
- 9. Basser PJ. Inferring microstructural features and the physiological state of tissues from diffusion-weighted images. NMR Biomed 1995;8:333–44 10.1002/nbm.1940080707 [DOI] [PubMed] [Google Scholar]
- 10. Flint JJ, Hansen B, Blackband SJ. Diffusion tensor microscopy data (15.6 μm in-plane) of white matter tracts in the human, pig, and rat spinal cord with corresponding tissue histology. Data Brief 2016;9:271–74 10.1016/j.dib.2016.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flint JJ, Hansen B, Fey M, et al. . Cellular-level diffusion tensor microscopy and fiber tracking in mammalian nervous tissue with direct histological correlation. Neuroimage 2010;52:556–61 10.1016/j.neuroimage.2010.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hansen B, Flint JJ, Heon-Lee C, et al. . Diffusion tensor microscopy in human nervous tissue with quantitative correlation based on direct histological comparison. Neuroimage 2011;57:1458–65 10.1016/j.neuroimage.2011.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beaulieu C. The basis of anisotropic water diffusion in the nervous system: a technical review. NMR Biomed 2002;15:435–55 10.1002/nbm.782 [DOI] [PubMed] [Google Scholar]
- 14. Rosenkrantz AB, Padhani AR, Chenevert TL, et al. . Body diffusion kurtosis imaging: basic principles, applications, and considerations for clinical practice. J Magn Reson Imaging 2015;42:1190–202 10.1002/jmri.24985 [DOI] [PubMed] [Google Scholar]
- 15. Kjølby BF, Khan AR, Chuhutin A, et al. . Fast diffusion kurtosis imaging of fibrotic mouse kidneys. NMR Biomed 2016;29:1709–19 10.1002/nbm.3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fieremans E, Jensen JH, Helpern JA. White matter characterization with diffusional kurtosis imaging. Neuroimage 2011;58:177–88 10.1016/j.neuroimage.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hansen B, Jespersen SN. Kurtosis fractional anisotropy, its contrast and estimation by proxy. Sci Rep 2016;6:2399 10.1038/srep23999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hansen B, Lund TE, Sangill R, et al. . Experimentally and computationally fast method for estimation of a mean kurtosis. Magn Reson Med 2013;69:1754–60 10.1002/mrm.24743 [DOI] [PubMed] [Google Scholar]
- 19. Hansen B, Jespersen SN. Recent developments in fast kurtosis imaging. Front Phys 2017;5 10.3389/fphy.2017.00040 [DOI] [Google Scholar]
- 20. Garza-Villarreal EA, Chakravarty MM, Hansen B, et al. . The effect of crack cocaine addiction and age on the microstructure and morphology of the human striatum and thalamus using shape analysis and fast diffusion kurtosis imaging. Transl Psychiatry 2017;7:e1122 10.1038/tp.2017.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hui ES, Fieremans E, Jensen JH, et al. . Stroke assessment with diffusional kurtosis imaging. Stroke 2012;43:2968–73 10.1161/STROKEAHA.112.657742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weber RA, Hui ES, Jensen JH, et al. . Diffusional kurtosis and diffusion tensor imaging reveal different time-sensitive stroke-induced microstructural changes. Stroke 2015;46:545–50 10.1161/STROKEAHA.114.006782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bay V, Kjolby BF, Iversen NK, et al. . Stroke infarct volume estimation in fixed tissue: comparison of diffusion kurtosis imaging to diffusion weighted imaging and histology in a rodent MCAO model. PLos One 2018;13:e019616 10.1371/journal.pone.0196161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Falangola MF, Jensen JH, Tabesh A, et al. . Non-Gaussian diffusion MRI assessment of brain microstructure in mild cognitive impairment and Alzheimer's disease. Magn Reson Imaging 2013;31:840–46 10.1016/j.mri.2013.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inglese M, Bester M. Diffusion imaging in multiple sclerosis: research and clinical implications. NMR Biomed 2010;23:865–72 10.1002/nbm.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arab A, Ruda-Kucerova J, Minsterova A, et al. . Diffusion kurtosis imaging detects microstructural changes in a methamphetamine-induced mouse model of Parkinson's disease. Neurotox Res 2019. June 18. [Epub ahead of print] 10.1007/s12640-019-00068-0 [DOI] [PubMed] [Google Scholar]
- 27. Khairnar A, Latta P, Drazanova E, et al. . Diffusion kurtosis imaging detects microstructural alterations in brain of alpha-synuclein overexpressing transgenic mouse model of parkinson's disease: a pilot study. Neurotox Res 2015;28:281–89 10.1007/s12640-015-9537-9 [DOI] [PubMed] [Google Scholar]
- 28. Wang JJ, Lin WY, Lu CS, et al. . Parkinson disease: diagnostic utility of diffusion kurtosis imaging. Radiology 2011;261:210–17 10.1148/radiol.11102277 [DOI] [PubMed] [Google Scholar]
- 29. Zhang G, Zhang Y, Zhang C, et al. . Diffusion kurtosis imaging of substantia nigra is a sensitive method for early diagnosis and disease evaluation in Parkinson's disease. Parkinsons Dis 2015;2015:207624 10.1155/2015/207624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Cauter S, Veraart J, Sijbers J, et al. . Gliomas: diffusion kurtosis MR imaging in grading. Radiology 2012;263:492–501 10.1148/radiol.12110927 [DOI] [PubMed] [Google Scholar]
- 31. Bai Y, Lin Y, Tian J, et al. . Grading of gliomas by using monoexponential, biexponential, and stretched exponential diffusion-weighted MR imaging and diffusion kurtosis MR imaging. Radiology 2016;278:496–504 10.1148/radiol.2015142173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grossman EJ, Ge Y, Jensen JH, et al. . Thalamus and cognitive impairment in mild traumatic brain injury: a diffusional kurtosis imaging study. J Neurotrauma 2012;29:2318–27 10.1089/neu.2011.1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grossman EJ, Jensen JH, Babb JS, et al. . Cognitive impairment in mild traumatic brain injury: a longitudinal diffusional kurtosis and perfusion imaging study. AJNR Am J Neuroradiol 2013;34:951–57 10.3174/ajnr.A3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Næss-Schmidt E, Blicher JU, Eskildsen SF, et al. . Microstructural changes in the thalamus after mild traumatic brain injury: a longitudinal diffusion and mean kurtosis tensor MRI study. Brain Injury 2017;31:230–36 10.1080/02699052.2016.1229034 [DOI] [PubMed] [Google Scholar]
- 35. Østergaard L, Engedal TS, Aamand R, et al. . Capillary transit time heterogeneity and flow-metabolism coupling after traumatic brain injury. J Cereb Blood Flow Metab 2014;34:1585–98 10.1038/jcbfm.2014.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Næss-Schmidt E, Blicher JU, Tietze A, et al. . Diffusion MRI findings in patients with extensive and minimal post-concussion symptoms after mTBI and healthy controls: a cross sectional study. Brain Inj 2018;32:91–8 10.1080/02699052.2017.1377352 [DOI] [PubMed] [Google Scholar]
- 37. Poot DH, den Dekker AJ, Achten E, et al. . Optimal experimental design for diffusion kurtosis imaging. IEEE Trans Med Imaging 2010;29:819–29 10.1109/TMI.2009.2037915 [DOI] [PubMed] [Google Scholar]
- 38. Hansen B, Lund TE, Sangill R, et al. . Experimental considerations for fast kurtosis imaging. Magn Reson Med 2016;76:1455–68 10.1002/mrm.26055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hansen B, Lund TE, Sangill R, et al. . Erratum: Hansen, Lund, Sangill, and Jespersen. Experimentally and computationally fast method for estimation of a mean kurtosis. Mag Reson Med 2014;71:2250. [DOI] [PubMed] [Google Scholar]
- 40. Glenn GR, Helpern JA, Tabesh A, et al. . Quantitative assessment of diffusional kurtosis anisotropy. NMR Biomed 2015;28:448–59 10.1002/nbm.3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alstrup AKO, Munk OL, Jensen TH, et al. . Magnetic resonance imaging and computed tomography as tools for the investigation of sperm whale (Physeter macrocephalus) teeth and eye. Acta Vet Scand 2017;59:38 10.1186/s13028-017-0307-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hansen B, Jespersen SN. Data for evaluation of fast kurtosis strategies, b-value optimization and exploration of diffusion MRI contrast. Sci Data 2016;3:160072 10.1038/sdata.2016.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hansen B, Khan AR, Shemesh N, et al. . White matter biomarkers from fast protocols using axially symmetric diffusion kurtosis imaging. NMR Biomed 2017;30 10.1002/nbm.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hansen B, Shemesh N, Jespersen SN. Fast imaging of mean, axial and radial diffusion kurtosis. Neuroimage 2016;142:381–93 10.1016/j.neuroimage.2016.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Setsompop K, Fan Q, Stockmann J, et al. . High-resolution in vivo diffusion imaging of the human brain with generalized slice dithered enhanced resolution: simultaneous multislice (gSlider-SMS). Magn Reson Med 2018;79:141–51 10.1002/mrm.26653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khan AR, Chuhutin A, Wiborg O, et al. . Biophysical modeling of high field diffusion MRI demonstrates microstructural aberration in chronic mild stress rat brain. Neuroimage 2016;142:421–30 10.1016/j.neuroimage.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Khan AR, Hansen B, Wiborg O, et al. . Diffusion MRI and MR spectroscopy reveal microstructural and metabolic brain alterations in chronic mild stress exposed rats: a CMS recovery study. Neuroimage 2018;167:342–53 10.1016/j.neuroimage.2017.11.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lasič S, Szczepankiewicz F, Eriksson S, et al. . Microanisotropy imaging: quantification of microscopic diffusion anisotropy and orientational order parameter by diffusion MRI with magic-angle spinning of the q-vector. Front Phys 2014;2 10.3389/fphy.2014.00011 [DOI] [Google Scholar]
- 49. Avram AV, Ozarslan E, Sarlls JE, et al. . In vivo detection of microscopic anisotropy using quadruple pulsed-field gradient (qPFG) diffusion MRI on a clinical scanner. Neuroimage 2013;64:229–39 10.1016/j.neuroimage.2012.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]