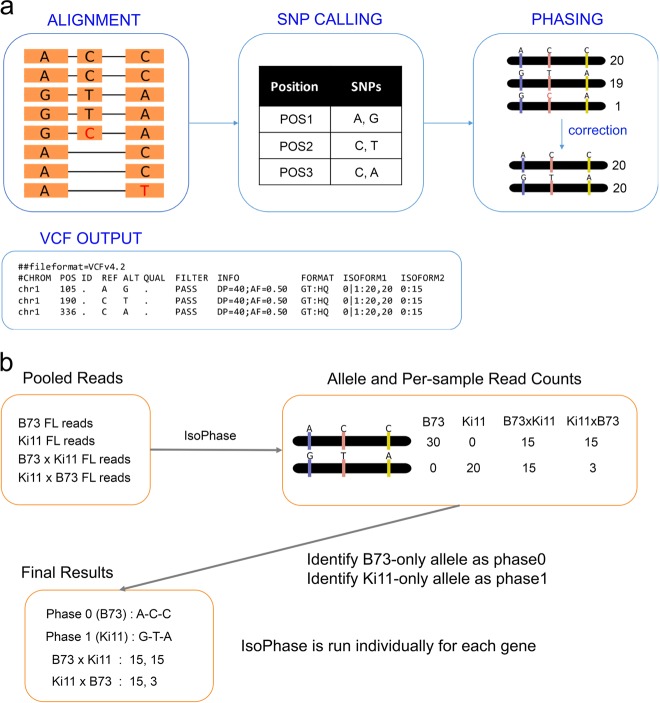
Fig. 3. IsoPhase workflow.
a For each gene, full-length reads from all 12 samples are aligned to a gene region. SNPs are called individually for each position using Fisher’s exact test with the Bonferroni correction, applied with a p-value cutoff of 0.01. Only substitution SNPs are called. The full-length reads are then used to reconstruct the haplotypes, and a simple error-correction scheme is applied to obtain the two alleles. b To determine which allele is derived from B73 vs. Ki11, we use the FL count information associate with the homozygous parents: B73 would only express the B73 allele, whereas Ki11 would only express the Ki11 allele.