Figure 6.
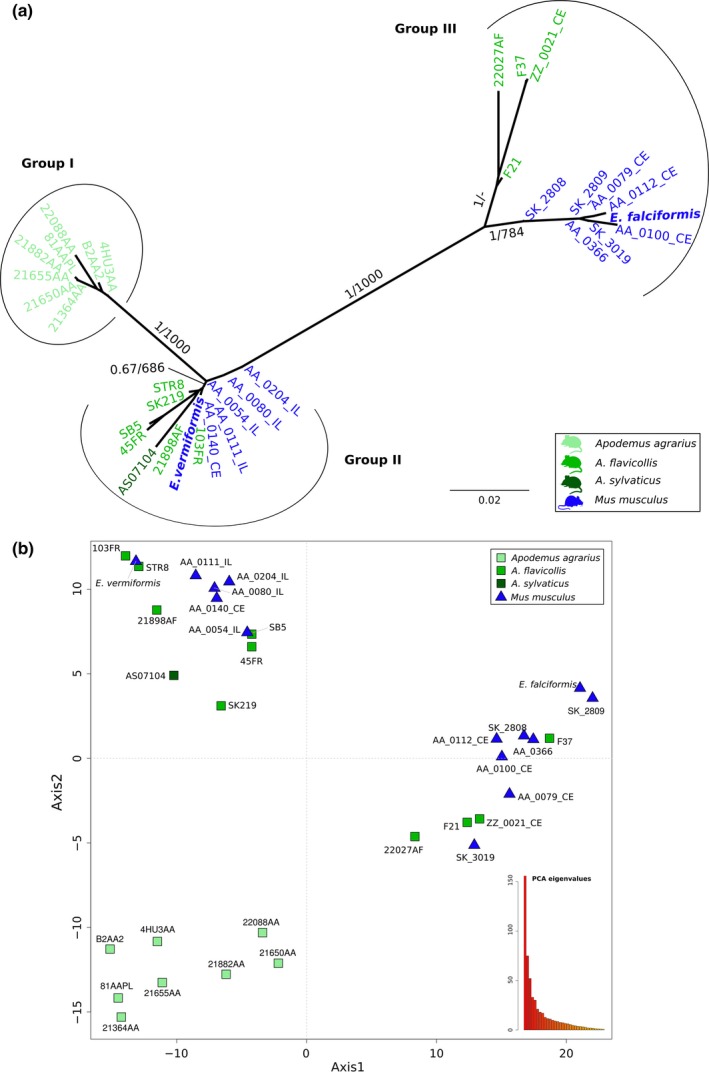
Nuclear multilocus genotyping of Eimeria isolates from Mus musculus and Apodemus. (a) The phylogenetic tree was estimated with a multimarker dataset formed with 35 nuclear markers from 31 Eimeria isolates derived from wild Mus musculus and three species of Apodemus (A. agrarius, A. sylvaticus, A. flavicollis). Eimeria falciformis and E. vermiformis sequences were included as reference. The scale bar represents sequence divergence. Color represents the host of origin for the isolates. Bootstrap support values and Bayesian posterior probabilities are shown on branches. (b) Principal component analysis based on single nucleotide polymorphisms (SNPs) from the same Eimeria isolates. Samples form three clusters. Shape indicates the genus of host and colors the species. Eigenvalues of the dimensions are shown in an insert to visualize the proportion of variance explained by the axes
