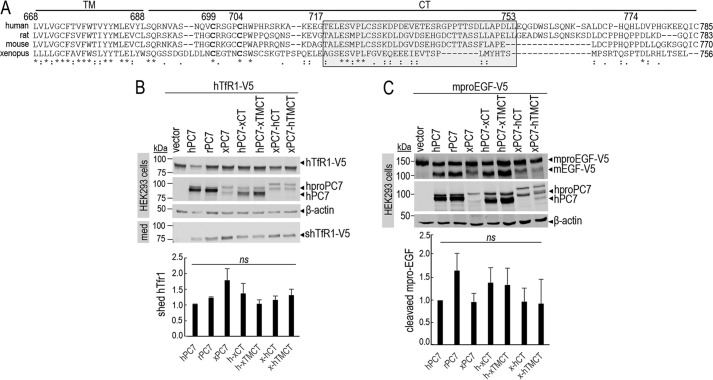
Figure 1.
Common critical residues in the CT of PC7 of different species. A, sequence alignment of PC7 from different species: human, rat, mouse, and X. laevis. The TM and the CT are indicated by two different horizontal lines. Conserved amino acids between species are noted with stars (identical) and either single dots (similar) or double dots (similar functionality). B, Western blot analysis of cell lysates (cells) and media (med) after overexpression of hTfR1-V5 with empty vector, human PC7 (hPC7), rat PC7 (rPC7), Xenopus PC7 (xPC7) or its chimeric forms (Xenopus/human transmembrane domain and cytosolic tail (xTMCT/hTMCT) and Xenopus/human cytosolic tail (xCT/hCT)) in HEK293 cells. Normalization was performed using the hTfR1-V5 shed by hPC7 as a reference. These results are representative of three independent experiments. C, Western blot analysis of cell lysates after overexpression of mouse proEGF (mproEGF) with empty vector, hPC7, rPC7, xPC7, or its truncated forms (Xenopus/human transmembrane domain + cytosolic tail (xTMCT/hTMCT) and Xenopus/human cytosolic tail (xCT/hCT)) in HEK293 cells. Normalization was performed using the mproEGF-V5 cleavage by hPC7 as a reference in HEK293 cells. These results are representative of three independent experiments. Error bars indicate averaged values ± S.D. ns, not significant (Student's t test).