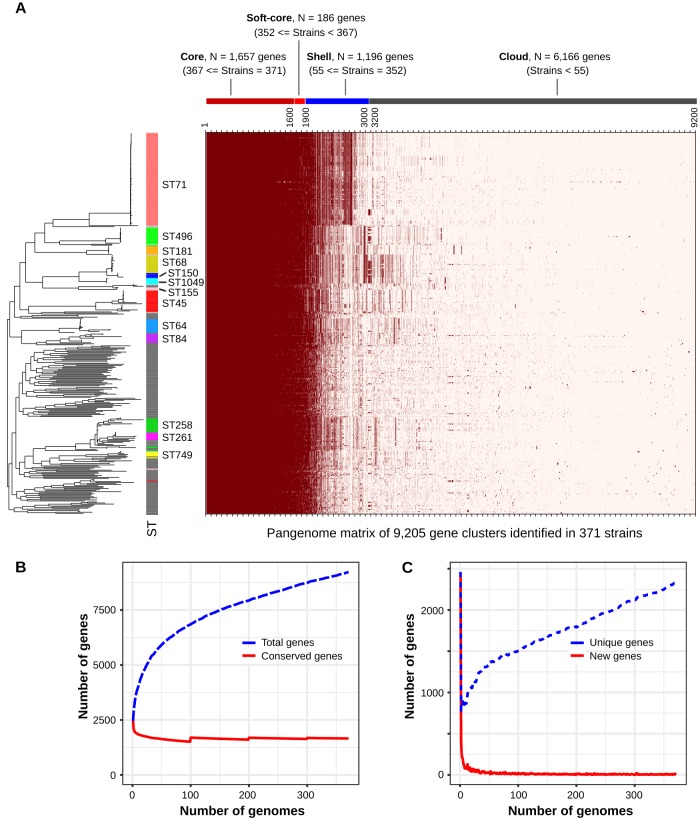
FIG 4.
Pangenome analysis of S. pseudintermedius. (A) Pangenome matrix showing the presence and absence of gene clusters in all 371 genomes. The average number of genes per isolate was ∼2,515 (median = 2,502), of which ∼1,843 were core genes (combining both core [N = 1,657] and soft core [N = 186]). The pangenome size of S. pseudintermedius was estimated to be ∼9,205 (total number of genes identified combining all 371 isolates). The rows in the heat map correspond to isolates, and columns correspond to orthologous gene clusters. The dark red color means a gene is present in that isolate, whereas a light color denotes absence of the gene. The rows in the heat map are ordered according to the whole-genome ML phylogeny plotted with the heat map. (B) Rarefaction curves showing the estimated core and pangenome sizes of S. pseudintermedius by ROARY. The blue and red lines show the change in the number of total genes (pangenome size) and conserved genes (core genes), respectively, as new genomes are added in the analysis. (C) Rarefaction curves showing the trend of unique (blue) and new (red line) genes being discovered as new genomes are added in the analysis. As shown, the S. pseudintermedius pangenome is still open, as both pangenome size and the number of unique genes being discovered continue to increase after all 371 genomes have been added.