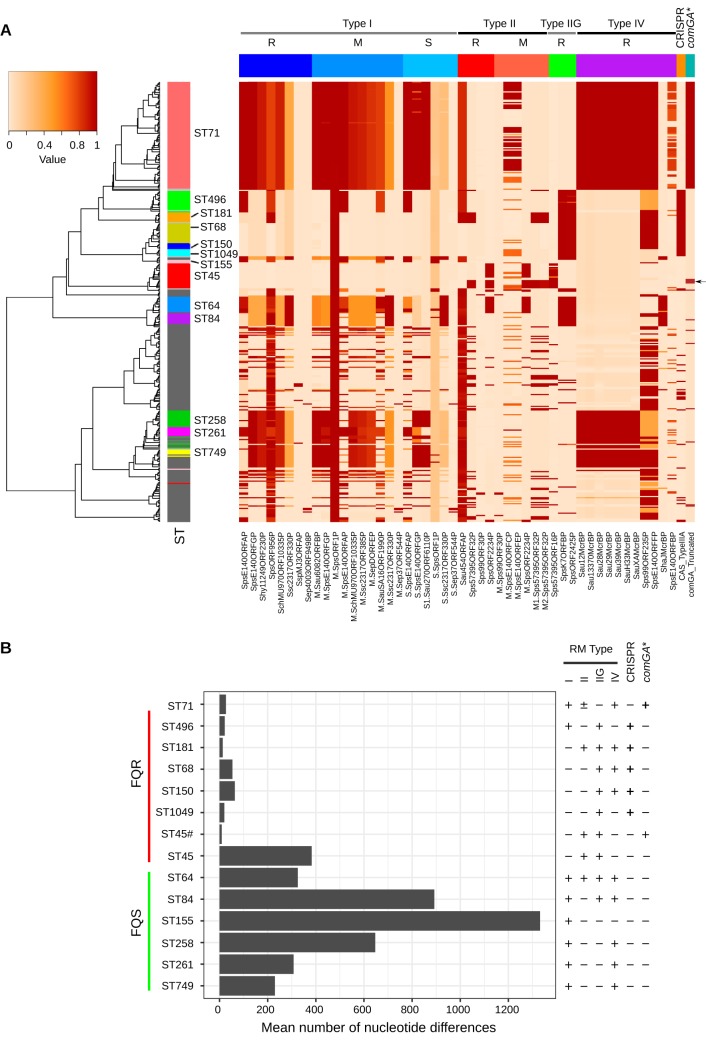
FIG 9.
Genetic barriers to HGT in S. pseudintermedius. (A) A heat map showing the distribution of RM type I, type II, type IIG, type IV, CRISPR/Cas, and disrupted comG (comGA*) systems in 371 genomes. The rows in the heat map correspond to isolates, and columns correspond to genes. The restriction (R), modification (M), and specificity (S) subunits of RM types are indicated. The colors in the heat map are based on the BSR value (range 0 to 1) obtained by the LS-BSR analysis, with a darker color corresponding to gene presence, and a lighter color corresponding to gene absence. Four ST45 isolates with disrupted comG are indicated with the arrow. (B) Bar graph showing core genomic diversity (mean number of nucleotide difference across the core genome) within major lineages. The presence (+) and absence (−) of RM types, CRISPR/Cas, and disrupted comG are shown with each lineage. The clones containing CRISPR/Cas or disrupted comG exhibited extremely reduced genetic diversity compared to those lacking these systems. ST45#, four ST45 isolates with disrupted comG; ST45, remaining 14 ST45 isolates with intact comG. Please refer to Table S5 for recombination analysis of these clones.