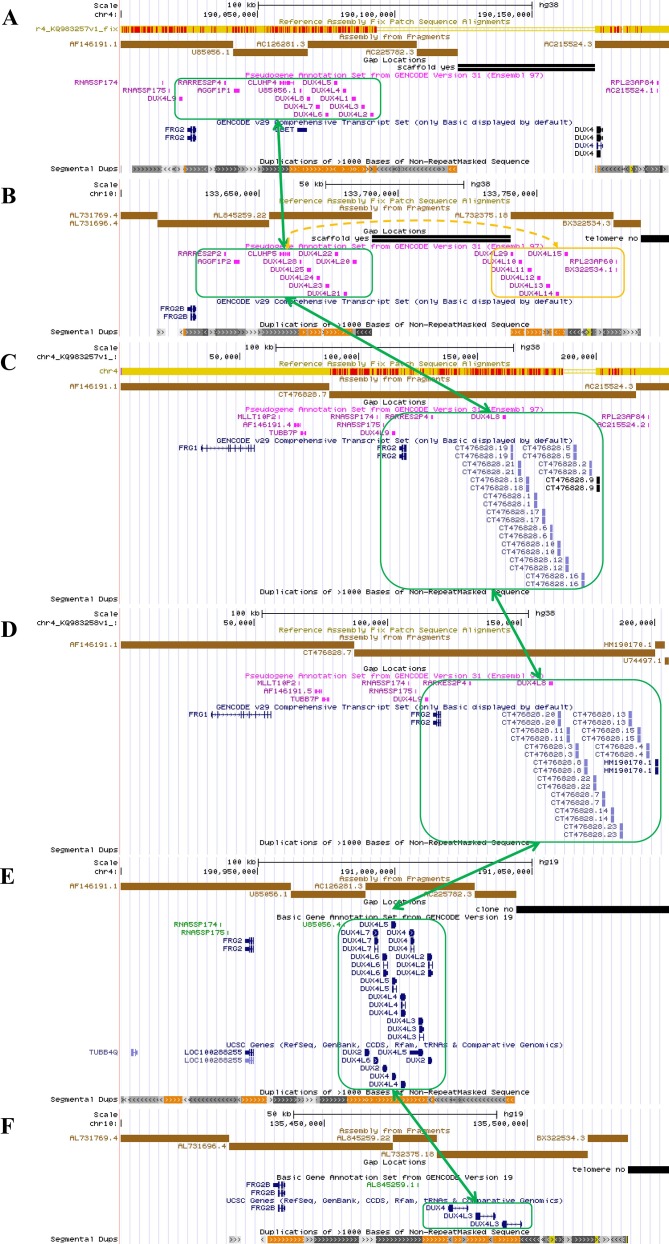
Figure 1.
An overview of the genomic architecture of segmental duplications at the chromosome 4q35 region and the 10q26 region. In the GRCh38 reference genome, 4q35 incorrectly shows two D4Z4 arrays (8 units 4qB type and 1.5 unit 4qA type) with a 50 kb gap between AC225782.3 (4qB) and AC215524.3 (4qA) (panel A). 10q26 incorrectly shows two D4Z4 arrays (each with 7 D4Z4 repeat units) with a 50 kb gap between AL845259.22 (10qA) and AL7323751.8 (10qA) (panel B). Two new patch scaffold sequences were recently added in CRCh38 patch 7 with 4qA configuration (KQ983257.1) without gap and with 4qA-L configuration (KQ983258.1) without gap, and we additionally illustrated them (panel C and D). The segmental duplication (green boxes) in 4q35 has high sequence identity with the corresponding region in 10q26, KQ983257.1 and KQ983258.1, while the distal D4Z4 array separated by the gap in 10q26 is marked with an orange box. The incorrect assembly gap is not present in 4q35 (panel E) or 10q26 (panel F) in the GRCh37 genome assembly.