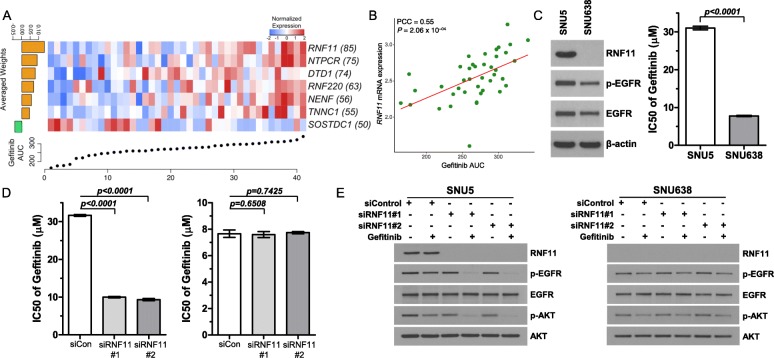
Fig. 4.
Transcriptome correlates of gefitinib sensitivity. a Elastic-net regression results of transcriptome features that predict pharmacological response to gefitinib. The bottom scatter plot represents drug response for gefitinib-treated tumors. The upper heatmap shows the top extracted features in the model. The left bar graph shows the averaged weight of each predictive feature. The number of appearances in 100 bootstraps is indicated in parentheses. b Scatter plot revealing a linear correlation between gefitinib AUC and RNF11 transcriptome expression. Correlation coefficients and P values were obtained by Pearson correlation analysis. c Immunoblot analysis of RNF11, p-EGFR, EGFR in gastric cancer cell-lines. β-Actin was used as a loading control (left panel). Cell proliferation assay in EGFR-activated gastric cancer cell-lines (right panel). Cancer cells were exposed to gefitinib for 72 h, and then, cell viability was measured. d Gastric cancer cell-lines with high (SNU5; left panel) and low (SNU638; right panel) RNF11 expression were transiently transfected with 10 nM of siRNF11 and treated with gefitinib for 72 h the next day. The results are represented as the mean ± SD of triplicate wells and are representative of three independent experiments. e Immunoblot analysis of EGFR signaling-related molecules, including p-EGFR, EGFR, p-AKT, and AKT in gastric cancer cell-lines that were transiently transfected with 10 nM of siRNF11 and treated with gefitinib for 4 h the next day. P values in c, d were derived from two-sided Student’s t tests