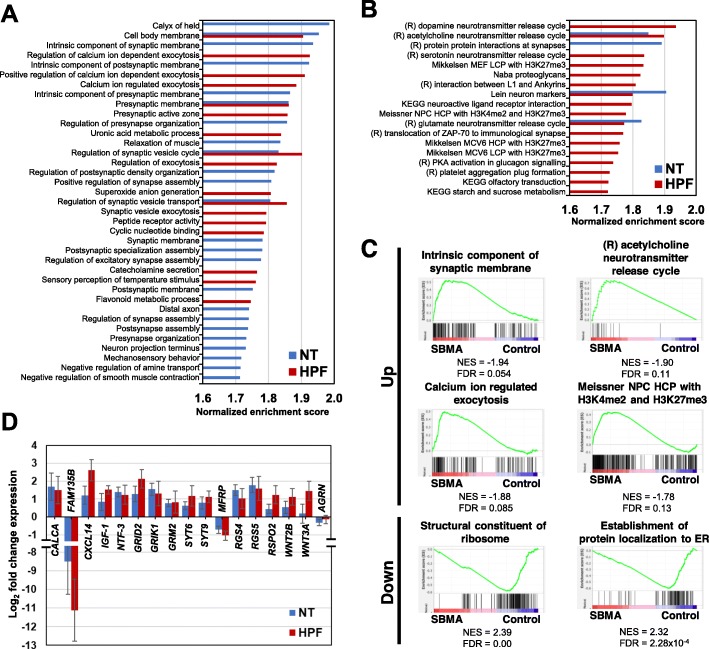
Fig. 6.
Enrichment of synapse-related pathology by GSEA. a Gene ontology (GO) enrichment analysis. Up-regulated gene sets in SBMA with FDR < 0.2 are shown. Blue indicates enriched in NT, and red in HPF. For negatively enriched GO terms, see also Tables S4 and S5, Additional file 3. b Pathways significantly enriched in SBMA with FDR < 0.2 are shown. Blue indicates enriched in NT, and red in HPF. (R), Reactome is a curated database of pathways and reactions in human biology. For negatively enriched pathway terms, see also Tables S6 and S7, Additional file 3. c Representative enrichment plots. Synapse, neurotransmitter, exocytosis, and epigenetics associated gene sets were involved in enrichment of SBMA. Ribosome and ER were negatively enriched in SBMA. The green curves show the enrichment score and reflect the degree to each black line, which shows a position of a gene in the ranked list of genes. NES, normalized enrichment score. FDR, false discovery rate. ER, endoplasmic reticulum. d The focused gene expression from DEGs and GSEA. The vertical axis represented relative log2 fold change (SBMA / Control) in the gene expression (n = 4, mean ± SEM)