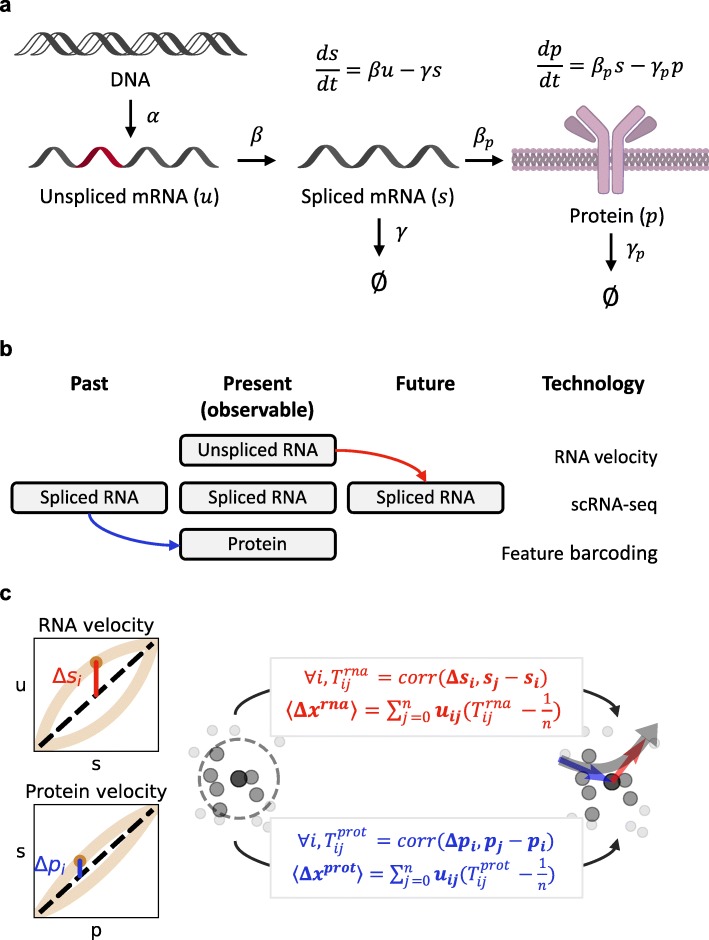
Fig. 1.
Model structure and parameter inference. a A single gene’s information transfer through transcription, splicing, and translation, and the ordinary differential equations governing the spliced mRNA and protein populations. b Conceptual framework for extrapolation from snapshot sequencing data. c Protein acceleration workflow: estimation of equilibrium states u = γs and s = γpp (black dashed lines) from imputed gene-specific population data (light brown), gene-specific extrapolation to calculate Δsi and Δpi, identification of nearest neighbors (dark gray: cell i, intermediate gray: n neighboring cells j, light gray: non-neighbor cells, circle: neighborhood), calculation of transition probabilities and embedded velocities (red: RNA velocity, blue: protein velocity, Tij: transition probability from cell i to neighbor j, uij: unit vector from cell i to neighbor j), and visualization of acceleration (blue arrow: protein velocity, red arrow: RNA velocity, combined curvature: gray Bézier curve)