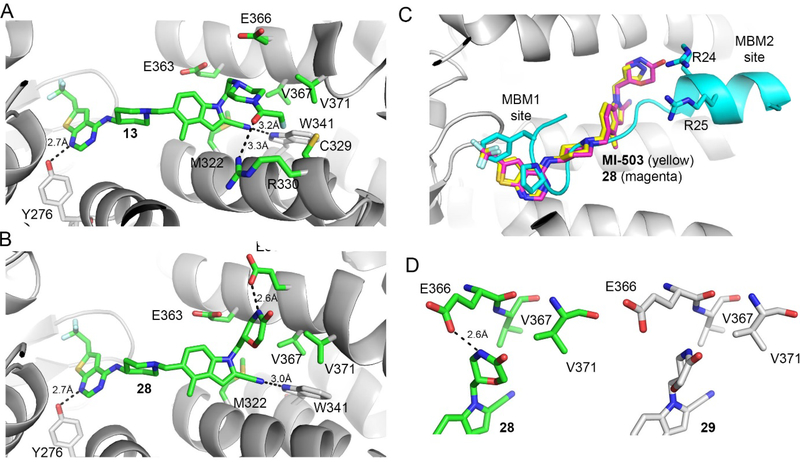
Figure 2. Structure-based optimization results in potent menin-MLL inhibitors.
a) Crystal structure of the menin-compound 13 complex (PDB code 6BXH). Residues on menin involved in the interactions with the piperazine ring and the terminal amide of 13 are shown in stick representation. Hydrogen bond with Arg330 is marked. b) Crystal structure of the menin-compound 28 complex (PDB code 6BXY). Residues on menin involved in the interactions with the morpholinone ring in 28 are shown in stick representation. Hydrogen bond with Glu366 is marked. c) Superposition of the crystal structures of menin in complex with MI-503, 28 and MLL peptide encompassing MBM1 and MBM2 motifs (3U88 in PDB). d) Comparison of the binding mode of 28 (left) and 29 (right, PDB code 6BY8) to menin.