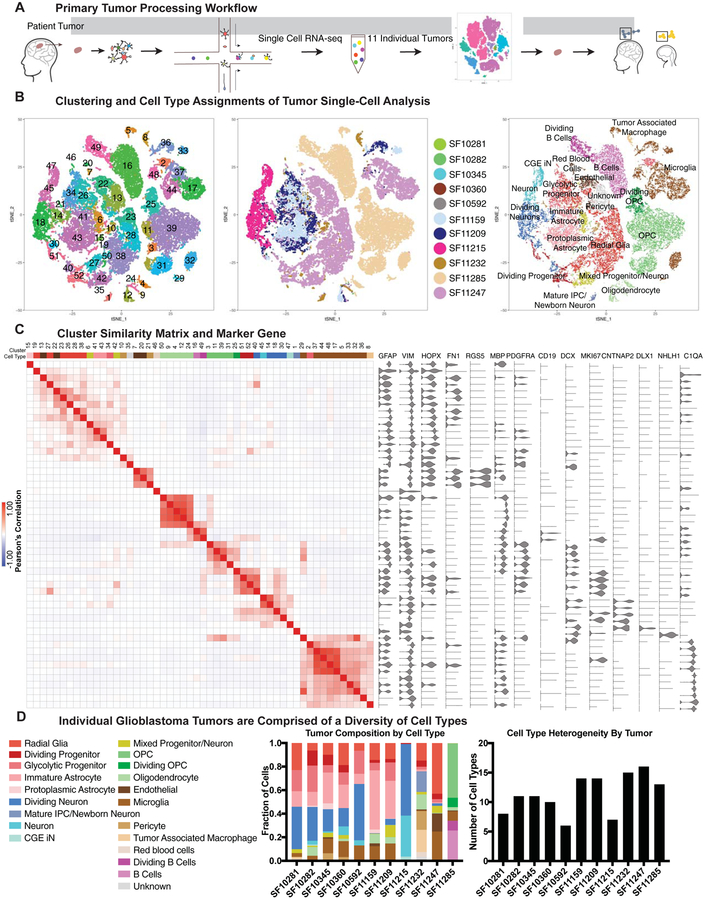
Figure 1. Single cell RNA-sequencing of primary glioblastoma tumors creates an atlas of tumor cell types.
a) Schematic of the workflow used to create the tumor atlas. Primary tumor resections are obtained and dissociated for single-cell sequencing. Clustering is performed and cell clusters are compared to annotated clusters from adult and developing human datasets. b) tSNE plots showing the clustering of primary tumor cells, colored by cluster, tumor of origin, and annotated cell type. c) Similarity matrix of clusters from primary glioblastoma analysis, correlated in the space of marker genes. Heatmap color indicates the Pearson’s correlation, and above the heatmap is the annotated cell type for each cluster. To the right of the heatmap are marker gene violin plots depicting the distribution of cell type genes across clusters. Some genes (such as GFAP) are broadly expressed but strongly enriched in clusters where the marker is associated with cell type (such as cluster 15, protoplasmic astrocyte-like cells). d) Composition of individual tumors is shown as a proportion of single-cells identified as each annotated cell type. Stacked barchart shows the proportional composition for each tumor, and graph to the right shows the number of cell types represented in each tumor.