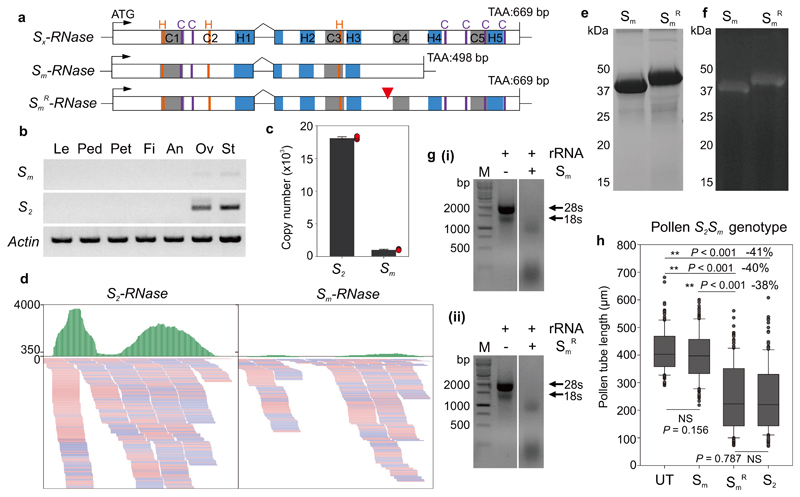
Figure 4. A truncated Sm-RNase appears to be responsible for the loss of SI in citrus.
a, Cartoon showing the gene structure of the Sx-RNase (top) and its mutant form (middle). Sx-RNase indicates the length and structure of the normal S-RNase with five conserved domains (C1-C5, grey boxes) and five hypervariable domains (H1-H5, blue boxes). Sm-RNase harbors a single nucleotide deletion resulting in a premature stop codon. The ‘recovered’ SmR-RNase (bottom) has a single nucleotide inserted (red triangle) that recovered the full length gene. Conserved histidine and cysteine residues are indicated in orange and purple, based on the deduced amino acid sequences. b, Expression of Sm- (Sm) and S2-RNase (S2) quantified using RT-PCR of different tissues from a S2Sm plant (Le: leaf; Ped: pedicel; Pet: petal; Fi: filament; An: anther; Ov: ovary; St: style). The Sm-RNase is expressed at a much lower level than the S2-RNase, in a tissue-specific manner (only in pistils: ovary and style tissues). c, Absolute copy number of the S2- and Sm-RNase (extrapolated from the equation in Supplementary Fig. 18). The copy number of Sm-RNase in 50 μg S2Sm style RNA is much lower than that of S2-RNase. Error bars are shown for mean copy number ± SEM (n = 3 biological replicates; red dots indicate individual samples). d, IGV tracks displaying sequencing read clusters of S2- and Sm-RNase gene from S2Sm style RNA-seq. The green bars depict the number of the reads mapped to the reference. The reads mapped to S2-RNase are clearly more than the reads mapped to Sm-RNase. Partial alignment of the RNA-mapping is shown below, with pink and blue representing the different read strands. e, SDS-PAGE analysis of recombinant Sm- (Sm) and SmR-RNase (SmR). The Mr of the fusion protein Sm-RNase-GST (39 kDa) is lower than the “restored” SmR-RNase-GST (45 kDa). f, RNase-activity gel of the recombinant Sm- (Sm) and SmR-RNase (SmR). The mutated Sm-RNase and the “recovered” SmR-RNase have similar RNase activities. g, The recombinant Sm- (i) and SmR-RNase (ii) both degrade citrus 28S and 18S rRNA (agarose gel assay). Experiments were repeated independently twice times for panel b and three times for panels e and g, with the similar results for each experiments. h, The SmR-RNase recombinant protein displays inhibitory activity against pollen. Box plots show the distribution of individual pollen tube lengths in an in vitro bioassay of recombinant Sm-, SmR- and S2 RNase against pollen from a plant with S2Sm genotype. The Sm-RNase did not significantly (NS) inhibit pollen tubes from a S2Sm plant when compared against untreated (UT) pollen; the SmR-RNase exhibited significant inhibitory activity (**) reducing the length of pollen tubes by ~40% and was not significantly different (NS) from that of the S2-RNase. The length of >50 pollen tubes was measured for each replicate (n = 3 biologically independent replicates, >150 in total). One-way ANOVA analysis was used to compare the pollen tube lengths (treatment vs untreated control). The elements in box and whisker plots are the same as in Fig 2d.