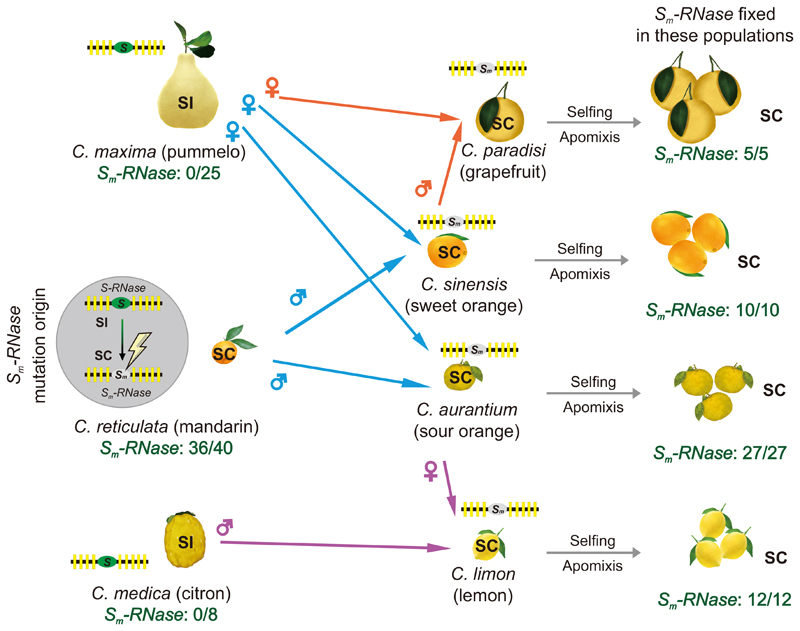
Figure 5. Postulated spread of the Sm-RNase and SC in Citrus.
Left hand side: Pummelo (top) is SI (no Sm-RNases were identified in 25 accessions). Citron (bottom) had a previously uncharacterized reproduction strategy, and we propose it is SI as we identified a single S-locus with one S-RNase and ~nine SLFs (and no Sm-RNases detected) in 8 accessions. We show a cartoon of the S-locus, with one S-RNase (green ellipse) and ~nine SLFs (yellow rectangles) by each citrus to indicate the status of its S-locus. Mandarin (middle) is SC. We observed the mutant Sm-RNase in 36/40 of mandarin accessions examined, and postulate that the mutant Sm-RNase arose spontaneously as an ancient event in wild mandarin (indicated by the lightning strike symbol) which converted mandarin from SI to SC (indicated by the grey ellipse). Middle: We propose that the Sm-RNase was subsequently transferred to other citrus species through pollination. The coloured arrows indicate the crosses between the parental genotypes proposed by Wu et al34 likely responsible for the origin of these citrus accessions: sweet orange and sour orange originated from crosses between pummelo and mandarin, with both acquiring SC from mandarin (blue arrows); grapefruit originated from a cross between sweet orange and pummelo, acquiring SC from sweet orange (orange arrows) and lemon acquired SC from a cross between sour orange and citron (purple arrows). Right hand side: Here we indicate that the Sm-RNase is responsible for the SC phenotype in grapefruit, sweet orange, sour orange and lemon, and that it is fixed in these populations, with all accessions examined being SC and containing the Sm-RNase (5/5, 10/10, 27/27, 12/12 respectively); none are SI. We propose that selfing and apomixis allowed the Sm-RNase to become fixed in these populations.