Populus euphratica WRKY1 increases the expression of the plasma membrane H+-ATPase gene by binding the promoter of PeHA1 in the presence of salt stress.
Keywords: DNA affinity purification sequencing, electrophoretic mobility shift assay, H+-ATPase, luciferase reporter assay, NaCl, non-invasive microtest technique, PeWRKY1, virus-induced gene silencing, W-box
Abstract
Plasma membrane proton pumps play a crucial role in maintaining ionic homeostasis in salt-resistant Populus euphratica under saline conditions. High levels of NaCl (200 mM) induced PeHA1 expression in P. euphratica roots and leaves. We isolated a 2022 bp promoter fragment upstream of the translational start of PeHA1 from P. euphratica. The promoter–reporter construct PeHA1-pro::GUS was transferred to tobacco plants, demonstrating that β-glucuronidase activities increased in root, leaf, and stem tissues under salt stress. DNA affinity purification sequencing revealed that PeWRKY1 protein targeted the PeHA1 gene. We assessed the salt-induced transcriptional response of PeWRKY1 and its interaction with PeHA1 in P. euphratica. PeWRKY1 binding to the PeHA1 W-box in the promoter region was verified by a yeast one-hybrid assay, EMSA, luciferase reporter assay, and virus-induced gene silencing. Transgenic tobacco plants overexpressing PeWRKY1 had improved expression of NtHA4, which has a cis-acting W-box in the regulatory region, and improved H+ pumping activity in both in vivo and in vitro assays. We conclude that salt stress up-regulated PeHA1 transcription due to the binding of PeWRKY1 to the W-box in the promoter region of PeHA1. Thus, we conclude that enhanced H+ pumping activity enabled salt-stressed plants to retain Na+ homeostasis.
Introduction
Salinity and secondary soil salinization cause serious ecological and environmental problems worldwide (Pitman and Läuchli, 2002; Hasegawa, 2013). For example, perennial trees, such as Populus euphratica, have to undergo long-term ionic stress in salt-affected soils. Populus euphratica is a model species for assessing the physiological mechanisms underlying salt tolerance in woody plants (Chen and Polle, 2010; Chen et al., 2014; Polle and Chen, 2015). Its ability to retain ionic homeostasis under salt stress is mostly related to H+-ATPases in the plasma membrane (PM) (Sun et al., 2009a, b). Proton pumps in P. euphratica have been shown to promote Na+ extrusion via Na+/H+ antiport across the PM (Sun et al., 2009a, b, 2010a, b, 2012). In addition to rapid post-translational modulations, particularly by the 14-3-3 protein (Palmgren et al., 1991; Svennelid et al., 1999; Camoni et al., 2000) and the negative modulator PKS5 kinase (Yang et al., 2010), PM H+-ATPases may undergo transcriptional regulation under salt stress (Gévaudant et al., 2007; Zhang et al., 2017).
Up-regulation of the PM H+-ATPase gene enabled Aeluropus littoralis to cope with salt stress (Olfatmiri et al., 2014). Constitutively expressed wild-type (WT) PM H+-ATPase 4 (wtPMA4) or a PMA4 mutant lacking the autoinhibitory domain enhances seed germination and plant growth under salt stress (Gévaudant et al., 2007). Ectopic expression of PeHA1, a gene encoding the PM H+-ATPase in P. euphratica, has been shown to significantly increase ATP hydrolysis and proton pumping activity in isolated PM vesicles from Arabidopsis (Wang et al., 2013). In addition, Song et al. (2017) demonstrated that GsJ11 overexpression increases transcription of H+-ATPase genes after alkaline treatment. We previously showed that PeJ3 overexpression suppressed AtPKS5 transcription in Arabidopsis. It is likely that PeJ3 enhances PM H+-ATPase activity by suppressing transcription of AtPKS5 in Arabidopsis (Zhang et al., 2017). However, the direct activation of PM H+-ATPase genes by transcription factors has not been thoroughly investigated in tree species under salt stress.
WRKY transcription factors constitute a large class of regulatory proteins in plants under adverse conditions (Eulgem et al., 2000; Rushton et al., 2010; Ishihama et al., 2011; Chen et al., 2012; Hu et al., 2013; Wang et al., 2014). Previous studies have demonstrated that WRKY transcription factors, such as Arabidopsis WRKY8 (AtWRKY8), AtWRKY18, AtWRKY40, and AtWRKY60, are involved in salt stress (Seki et al., 2002; Chen et al., 2010; Hu et al., 2013). AtWRKY8 is a positive regulator of salt stress (Hu et al., 2013). In transgenic wheat plants, TaWRKY79 enhances tolerance to salt stress (Qin et al., 2013). We have also shown that salinity induces WRKY1 in P. euphratica and that PeWRKY1 enables transgenic plants to extrude Na+ and retain K+ under NaCl stress (Shen et al., 2015a). The salt-induced Na+ efflux is thought to be promoted by H+-ATPase at the PM, as an inhibitor of H+ pumps (vanadate) significantly reduces the Na+ flux rate (Shen et al., 2015a).
The WRKY domain is defined by its N-terminal conserved amino acid sequence WRKYGQK. The WRKY protein specifically binds to the cis-acting element W-box, and its DNA sequence motif is (T)(T)TGAC(C/T) (Rushton et al., 1996, 2010; Eulgem et al., 2000). AtWRKY8 can bind to the W-boxes in the promoters of RD29A, RD29B, RD20, and ADH1 (Hu et al., 2013). We also found that the PeHA1 promoter regions harbored a cis-acting W-box (see Supplementary Fig. S1 at JXB online), implying that the transcription factor WRKY1 mediates PeHA1 expression.
In the present study, the objective was to investigate the interaction between PeWRKY1 and PeHA1 and its relevance to salt stress tolerance. Transcription factor binding protocols, such as DNA affinity purification sequencing (DAP-seq), yeast one-hybrid (Y1H) assay, EMSA, luciferase reporter assay (LRA), and virus-induced gene silencing (VIGS), were used to explore the binding of PeWRKY1 to the cis-acting W-box in the PeHA1 promoter. Salt-induced expression of the PeHA1 promoter was investigated in root and shoot tissues of transgenic tobacco. Furthermore, after overexpressing PeWRKY1 in tobacco plants, we examined in vivo and in vitro H+ pumping activities and Na+ fluxes under salt stress. The aim was to confirm whether the increased salt tolerance was due to PeWRKY1-up-regulated gene expression of H+-ATPase in transgenic plants.
Materials and methods
Plant material and salt treatment
One-year-old P. euphratica seedlings were obtained from the Xinjiang Uygur Autonomous Region of Northwest China. In April, seedlings were planted in 10 liter pots containing 1:1 sand and soil and kept at 20–25 °C in a greenhouse at Beijing Forestry University. Potted seedlings were irrigated and fertilized every 2 weeks with 1 liter of full-strength Hoagland’s nutrient solution. The photosynthetically active radiation (PAR) was 150–600 μmol m−2 s−1 during the culture period. The relative humidity was maintained at 40–70%. For salt treatment, 120 uniform seedlings were subjected to moderate salt stress by watering with 2 liters of NaCl (200 mM) in full-strength Hoagland’s nutrient solution. The salt-resistant P. euphratica did not exhibit symptoms of salt injury during salt exposure (Chen et al., 2001). Total RNA was extracted from the roots and upper mature leaves (leaf index numbers 4–20 from the shoot apex) at 0, 1, 3, 6, 12, 24, 48, and 72 h after salt treatment.
RNA isolation and cloning full-length PeWRKY1
Total RNA was extracted from leaves using the EASYspin Plus Complex Plant RNA Kit (Aidlab Biotech, Beijing, China) according to the manufacturer’s instructions. First-strand cDNA was synthesized in a reverse transcription reaction including 1 μg of total RNA, oligo(dT) primer, and M-MLV reverse transcriptase (Promega, Madison, WI, USA). PCR cloning of full-length PeWRKY1 was performed in a total volume of 25 μl containing 1 μl of cDNA product, 1 U of Ex Taq polymerase (Takara, Dalian, China), 2.5 μl of 10× Ex Taq buffer, 2 μl of dNTP mixture (2.5 mM), and 0.5 μl of forward and reverse primers (10 μM) designed based on the homologous WRKY1 sequence from P. euphratica. The 5′ to 3′ primer sequences were: forward, 5′-ATGGCTGCTTCTTCAGGGAG-3′; and reverse, 5′-CTACCAAAAACTCTCTACTTCC-3′. The PCR product was gel purified and then ligated to the pMD18-T vector (Takara) for DNA sequencing.
Quantitative real-time PCR analysis of PeWRKY1 and PeHA1 in P. euphratica
Total RNA was extracted from P. euphratica roots and upper leaves using an EASYspin Plus Complex Plant RNA Kit (Aidlab Biotech, Beijing, China) according to the manufacturer’s instructions. The P. euphratica leaves (leaf index numbers 4–20 from the shoot apex) and roots were sampled after 0, 1, 3, 6, 12, 24, 48, and 72 h of salt treatment (200 mM NaCl). The isolates then were treated with RNase-free DNase I (Takara) for 30 min at 37 °C. First-strand cDNA was synthesized using 1.0 μg of total RNA with the SuperScript™ First Strand Synthesis System (Promega). The cDNA product (1 μl) was used for SYBR Green-based real-time PCR (RT-PCR) analysis in the 7500 Fast real-time PCR system (Applied Biosystems, Carlsbad, CA, USA). Three biological replicates were established in each treatment group and PeACT7 was used as an endogenous reference. Three technical replicates were analyzed for each biological sample. Supplementary Table S1 lists the forward and reverse primers for PeHA1, PeWRKY1, and PeACT7. The expression of the target gene was normalized to that of the reference gene (PeACT7) using the 2−∆∆Ct method. The PCR running conditions were as follows: 95 °C for 5 min followed by 34 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and finally 72 °C for 10 min (Shen et al., 2015a, b).
DNA affinity purification sequencing
DAP-seq binding assays were performed as described by O’Malley et al. (2016) and Bartlett et al. (2017), with modifications as described briefly herein.
Nuclear isolation and genomic DNA exaction
Fresh P. euphratica leaves were ground to fine powder using liquid nitrogen. The powder was resuspended in 2 ml of cold nuclear isolation buffer. The solution was filtered through a 30 μm mesh cell strainer and centrifuged at 2000 g for 4 min at 4 °C, after which the supernatant was discarded. The nuclear DNA was resuspended in 100 μl of phosphate-buffered saline (PBS). Then 1 μl of RNase was added at 37 °C for 30 min, followed by 3 μl of proteinase K at 55 °C for 30 min, and 100 μl of 2× cetyltrimethylammonium bromide (CTAB) at 65 °C for 30 min. Finally 200 μl of phenol/chloroform/isoamyl alcohol was added and centrifuged at 11 000 g for 10 min at room temperature. The supernatant was transferred into a new 1.5 ml Eppendorf tube, to which 2.5× volumes of ethanol were added, and incubated at –80 °C for 1 h, followed by centrifuging at 11 000 g for 10 min at room temperature. The supernatant was discarded, and the tube was dried at room temperature. The DNA was dissolved in 50 μl of Tris-EDTA (TE) buffer.
DAP-seq genomic DNA library preparation
Genomic DNA (gDNA; 5 μg in 130 μl of TE buffer) was fragmented to an average of 200 bp using a Covaris M220 (Woburn, MA, USA) as per the manufacturer’s recommended settings. The fragmented gDNA then was purified using AMPure XP beads (Beckman Coulter, Inc., Indianoplis, IN, USA) at a DNA to beads ratio of 0.7–1.1. The beads were incubated with the gDNA for 5 min at room temperature, placed on a magnet to immobilize the beads, and then the supernatant was removed. The beads were washed twice with 200 μl of 80% ethanol and allowed to dry. Once dry, they were resuspended in 22 μl of resuspension buffer, incubated at room temperature for 5 min, and placed on the magnet. The DNA-containing supernatant was then transferred to a new tube. Libraries were constructed using the NEXTFLEX Rapid DNA-Seq Kit (PerkinElmer, Inc., Austin, TX, USA) according to the instruction manual.
DAP-seq protein expression
The coding sequencing of PeWRKY1 was cloned into a pFN19K HaloTag T7 SP6 Flexi expression vector. Halo–PeWRKY1 fusion protein was expressed using the TNT SP6 Coupled Wheat Germ Extract System (Promega) following the manufacturer’s specifications for expression in a 50 μl reaction with a 2 h incubation at 37°C. Expressed proteins were directly captured using Magne Halo Tag Beads (Promega).
DAP-seq binding assay and sequencing
The protein-bound beads were incubated with 50 ng of adaptor-ligated gDNA fragments on a rotator for 1 h at room temperature in 50 μl of wash/bind buffer. Beads were washed three times using the same wash buffer to remove unbound DNA fragments. The HaloTag beads were resuspended in 30 μl of elution buffer and heated to 98 °C for 10 min to denature the protein and release the bound DNA fragments into solution. The supernatant was transferred to a new well, and 25 μl were used in a 50 μl PCR employing the KAPA HiFi HotStart ReadyMixPCR Kit (Roche, Basel, Switzerland) for 10 cycles. PCR primers consisted of the full-length Illumina TruSeq Universal primer (5'–AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT–3') and an Illumina TruSeq Index primer (5'–CAAGCAGAAGACGGCATACGAGAT-NNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT–3'), where NNNNNN represents the 6 bp sequence index used for sample identification. The PCR product was purified and selected using AMPure XP beads (Beckman), as previously described, and resuspended in 20 μl of nuclease-free water. DNA concentrations were determined using a Qubit (Life Technologies, Burlington, Ontario, Canada). Eluted DNA fragments were sequenced on an Illumina NavoSeq. Negative control mock DAP-seq libraries were prepared without the addition of protein to the beads.
DAP-seq data processing
Reads were mapped to the P. euphratica genome sequence (https://www.ncbi.nlm.nih.gov/genome/13265; Ma et al., 2013) using BOWTIE2 (Langmead et al., 2012). Peak calling was done using Macs2 (Zhang et al., 2008). Association of DAP-seq peaks located upstream or downstream of the transcription start site (TSS) within 3.5 kb were analyzed using Homer (Heinz et al., 2010), based on the General Feature Format (GFF) files. Gene function annotation was blasted from NR, NT, Swissprot, and Pfam databases. FASTA sequences were obtained using BEDTools for motif analysis (Quinlan et al., 2010). Motif discovery was performed using MEME-Chip suite 5.0.5 (Machanick et al., 2011).
Luciferase reporter assay
The LRA was conducted according to previously described procedures (Hellens et al., 2005). Briefly, PeWRKY1 cDNAs were cloned into the effector plasmid pGreenII 62-SK, and the PeHA1 promoter (PeHA1pro, 1266 bp) sequence was cloned into the reporter plasmid pGreenII 0800-LUC. The negative controls were recombined without the PeWRKY1 or PeHA1 promoter. Agrobacterium tumefaciens GV3101 (pSoup-p19) was used for transformation of recombinant plasmids. Agrobacterium tumefaciens was inoculated into Luria–Bertani (LB) medium containing 25 μg ml−1 rifampicin and corresponding antibiotics (50 μg ml−1 kanamycin). The A. tumefaciens cultures were incubated at 28 °C in a shaker (250 rpm) until the optical density (OD600) reached 1.0. The bacterial solution was centrifuged at 4000 rpm for 5 min at room temperature, resuspended by tobacco injection buffer (10 mM MES pH 5.6, 10 mM MgCl2, 150 μM acetosyringone), and the OD600 was adjusted to 0.6. Equal concentrations and volumes of A. tumefaciens constructs, PeHA1pro-LUC, pGreenII 0800-LUC, PeWRKY1-62-SK, and pGreenII 62-SK were co-transformed into fully developed Nicotiana benthamiana leaves (6 weeks) using needle-free syringes, and incubated for 48–60 h (Chen et al., 2008). d-Fluorescein (1 mM) was sprayed onto the tobacco leaves and then photographed using an LB983 Night Owl II fluorescence imaging system (Berthold Technologies, Bad Wildbad, Germany). The relative luminescence intensity was calculated using Image-Pro Plus 6.0 (Media Cybernetics, Silver Spring, MD, USA). The LRA experiments were repeated at least three times.
Virus-induced gene silencing of PeWRKY1 in P. euphratica
VIGS assays were performed according to our previous reports (Shen et al., 2015a, b). Briefly, a 250 bp fragment of PeWRKY1 was cloned via PCR using the specific primers forward, 5′-CACAACTTCATCAGCCTT-3′ and reverse, 5′-AGACTGATACTGTGGTGGTTG-3′. Empty pTRV2, constructed pTRV2-PeWRKY1, and the pTRV1 vector were transformed into A. tumefaciens strain GV3101. Agrobacterium strains carrying pTRV2-PeWRKY1 and pTRV1, or empty pTRV2 and pTRV1 at a ratio of 1:1, infiltrated the mature leaves of P. euphratica. Agrobacterium-infiltrated seedlings were placed in a growth room in a greenhouse at Beijing Forestry University. A temperature of 28 °C was maintained during the light period and 18 °C during the dark period, with a 16 h photoperiod (07.00 h to 23.00 h). PAR was 150–200 μmol m−2 s−1. Relative humidity was maintained at 60–70%. After 45 d of Tobacco rattle virus (TRV) infection, seedlings were subjected to NaCl stress by top watering with 2 liters of 200 mM NaCl in full-strength Hoagland’s nutrient solution. RNA was extracted from upper mature leaves (leaf index numbers 4–20 from the shoot apex) after 6 h of salt treatment. The transcriptional levels of PeWRKY1 and PeHA1 were examined by quantitative RT-PCR as previously described. The housekeeping gene PeACT7 was used an internal reference, and three biological replicates were used for each RT-PCR.
Yeast one-hybrid assay
Using Gold Matchmaker™ (Clontech Laboratories, Inc., CA, USA), Y1H assays were used to verify the interaction between PeWRKY1 and the W-box motif of the PeHA1 promoter. The PeHA1 promoter was isolated via genomic walking with a series of PCR amplifications. Supplementary Table S2 shows the primer sequences used for PeHA1 promoter isolation. The promoter of PeHA1 (1266 bp) containing the W-box was cloned using specific primers and ligated into the pAbAi vector in front of the reporter gene AUR1-C [Aureobasidin A (AbA) resistance]. PeWRKY1 was cloned and expressed by fusion with the pGADT7 vector. The linearized PeHA1-pro-pAbAi plasmid was transferred into Y1H yeast, and the positive strain was identified by testing the AbA inhibition. The PeHA1-pro-pAbAi strain was plated on synthetic/–uracil (SD/-Ura) medium supplemented with 0, 700, and 800 ng ml−1 AbA. The Y1H strain was completely inhibited at 800 ng ml−1 AbA. Both positive yeast and Y1H blank strains were transferred to PeWRKY1-pGADT7, with an empty vector as the control, and cultured in SD/–leucine (-Leu) medium supplemented with 800 ng ml−1 AbA.
To further determine whether PeWRKY1 binds to the W-box or other regions of the promoter, the sequence of the W-box in the promoter of PeHA1 was mutated from TTGACC to TTGAAC. The Y1H assays were performed with the mutated W-box, as previously mentioned. Y1H assays were repeated three times, and representative results are shown.
EMSA
To construct the PeWRKY1–pGEX-4T-1 vector, the full-length coding sequence of PeWRKY1 was cloned with a glutathione S-transferase (GST) tag at the C-terminus of the pGEX-4T-1 vector. An empty pGEX-4T-1 vector was used as a negative control. The construct was transferred to Escherichia coli (BL21) for recombinant protein production. Briefly, the bacteria were cultured in LB medium at 37 °C until an OD600 of 0.5−0.6, and then cultured at 37 °C for 10 h after induction with 0.5 mM isopropyl-β-d-thiogalactoside (IPTG). The bacterial solution was collected by centrifugation, resuspended in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), and centrifuged after sonicating for 15 min. The supernatant of the cell lysate was collected and incubated with glutathione–Sepharose 4B beads (GE Healthcare, Buckinghamshire, UK) for 2 h at 4 °C, followed by washing five times with PBS. The beads were washed four times with 1 ml of GST elution buffer (50 mM Tris–HC1 pH 7.3, 10 mM reduced glutathione). The purified protein was collected in a 1.5 ml centrifuge tube. In parallel, 40 bp DNA fragments from the PeHA1 promoter (5′-CCCTATCGGGCAAGATTTGACCCGCCCGAGTTGATCCCTC-3′ and 5′-GAGGGATCAACTCGGGCGGGTCAAATCTTGCCCGATAGGG-3′, harboring the W-box motif) were biotin labeled at the 3′ end using an EMSA Probe Biotin Labeling Kit (Beyotime, Nantong, China). Unlabeled probes were subjected to cold competition experiments. Mutant W-box (TTGACC→TTAGCC) probes that were not labeled with biotin were used to confirm the binding specificity of the W-box with PeWRKY1. The cold probe concentrations were 5× and 10×, and the mutant probe was 5×. EMSA was performed using the Light Shift Chemiluminescent EMSA Kit (Beyotime) according to the manufacturer’s instructions. The GST protein was used as a negative control. The biotin signals were imaged using the ChemiDoc MP Imaging System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The EMSAs were repeated three times, and representative results are shown.
Generation of PeWRKY1 transgenic tobacco
To construct the 35S::PeWRKY1::pCABIA2300 overexpression vector, a 1788 bp fragment containing the PeWRKY1 ORF was amplified from P. euphratica cDNA using specific primers (forward, 5′-ATGGCTGCTTCTTCAGGGAG-3′; reverse, 5′-CTACCAAAAA CTCTCTACTTCC-3′) and then cloned into the pMD18-T vector. Next, the PeWRKY1 ORF was cloned into the EcoRI and BamHI sites of the pCABIA2300 vector driven by the 35S promoter. The 35S::PeWRKY1::pCABIA2300 construct was introduced into A. tumefaciens strain GV3101, and then transformed into WT N. tabacum using an Agrobacterium-mediated method (Shen et al., 2015a). The blank pCABIA2300 vector was introduced into WT tobacco plants as a control. Putative transgenic plants were selected on Murashige and Skoog (MS) medium containing 100 mg l−1 kanamycin and verified by semi-quantitative RT-PCR using PeWRKY1-specific primers (Supplementary Table S1; Supplementary Fig. S2; Shen et al., 2013). WT and transgenic tobacco plants were hydroponically cultured in a 1 liter container and placed in a growth chamber at 25±1 °C with a light intensity of 50 μmol m−2 s−1 and a photoperiod of 16 h light and 8 h dark. The relative humidity was maintained at 50–60%. Among the 15 independent transgenic lines, two independent transgenic lines (L-9 and L-12) exhibited higher abundance of PeWRKY1 transcript and were used for salt experiments. Supplementary Table S2 lists the specific primers used to clone the promoters of NtHA2 and NtHA4.
Phenotype tests of transgenic plants
Four-week-old seedlings of the WT, vector control (VC), and transgenic lines L-9 and L-12 were exposed to 0 or 200 mM NaCl for 3 d before measuring leaf fresh weight and malondialdehyde (MDA) content. Three individual plants per treatment were used for MDA and membrane potential measurements.
MDA content
Oxidative damage to lipids was estimated by measuring the amount of MDA in plants according to Wang et al. (2007), with modifications. The leaf samples from control and salt-stressed plants were mixed with 10% trichloroacetic acid (TCA) containing 0.6% 2-thiobarbituric acid (TBA) and cooled to room temperature after boiling for 15 min. The absorbance at 450, 532, and 600 nm was examined to calculate the MDA content (Wang et al., 2007).
Membrane potential measurements
WT, VC, and transgenic lines L-9 and L-12 were subjected to 0 or 200 mM NaCl for 24 h. The membrane potential at the apical region (300 μm or 400 μm from the root tip) was measured using silver/silver chloride microelectrodes [XY-CGQ03; Xuyue (Beijing) Sci. & Tech Co. Ltd, Beijing, China] as described previously (Sa et al., 2019).
Steady-state ion flux measurements
Four-week old seedlings of WT, VC, and PeWRKY1-transgenic L-9 and L-12 plants were treated with 0 or 200 mM NaCl for 24 h. The Na+ flux was recorded using a non-invasive microtest system (NMT-YG-100, Younger USA LLC, Amherst, MA, USA) with the ion flow rate measurement software iFluxes 1.0 (Younger USA) and signal processing software ASET 2.0 (Sciencewares, Falmouth, MA, USA), as previously described (Sun et al., 2009a, b, 2010a). Five to six individual plants of control and salinized plants were used, respectively, to examine the Na+ flux.
Detection of Na+ content in tobacco roots
Four-week old seedlings of WT, VC, L-9, and L-12 were transferred to MS liquid solution containing 0 or 200 mM NaCl for 24 h. The Na+ content was detected with a specific fluorescent probe (Zhang et al., 2017). The tobacco root was exposed to CoroNa-Green AM (20 µM) for 2 h in 5 mM MES-KCl loading buffer (pH 5.7) in the dark and then rinsed 4–5 times with MS solution. The fluorescent intensity was detected using a Leica SP5 confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany) at an excitation wavelength of 488 nm and emission wavelength of 510–530 nm. The relative fluorescence intensity was calculated using Image-Pro Plus 6.0 (Media Cybernetics). Three individual plants for each treatment were used for Na+ content measurements.
Purification of plasma membrane vesicles and H+-ATPase activity assays
Tobacco plants (WT, VC, L-9, and L-12) were homogenized in cold extraction lysis buffer [0.33 M sucrose, 0.2% (w/v) BSA, 5 mM EDTA, 5 mM DTT, 5 mM ascorbate, 1% (w/v) polyvinyl pyrrolidone (PVP-40), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 25 mM HEPES-KOH, pH 7.5]. The homogenate was filtered and centrifuged at 13 000 g for 10 min, then centrifuged for 1 h at 100 000 g. The resulting microsomal pellet was resuspended in buffer (0.33 M sucrose, 1 mM PMSF, and 5 mM potassium phosphate, pH 7.8) and added to a phase mixture of 6.3% (w/w) Dextran T-500 and 6.3% (w/w) polyethylene glycol 3350 in 5 mM potassium phosphate (pH 7.8), 0.33 M sucrose, and 3 mM KCl. The upper phase was collected, diluted with buffer containing 0.33 M sucrose, 1 mM PMSF, 2 mM DTT, and 10 mM MOPS-KOH (pH 7.5), and then centrifuged at 110 000 g for 30 min. The pellet was collected and used to measure H+-ATPase activity.
The H+-ATPase hydrolysis activity was measured according to protocols described by Wang et al. (2013). The reaction mixture (200 μl) contained 10 mM MOPS-KOH (pH 6.5), 50 mM KNO3, 50 mM KCl, 3 mM MgCl2, 1 mM NaN3, 0.5 mM Na2MoO4, and 20 mg of protein. The substrate, ATP (1 μl, 200 mM), was added to activate the reaction in the presence or absence of 400 μM Na3VO4. After 30 min of incubation at 37 °C, the reaction was stopped by adding 10% (w/v) TCA. H+-ATPase hydrolysis activity was measured by determining the Pi released from ATP.
The transport activity of PM H+-ATPase was measured by fluorescence quenching of acridine orange at 495 nm (Palmgren, 1990). The PM protein (50 mg ml−1) was mixed with reaction buffer containing 20 mM MES-KOH (pH 7.0), 140 mM KCl, 3 mM ATPNa2, 10 μM acridine orange, and 0.0125% (v/v) Triton X-100, and then incubated for 10 min at room temperature. The reaction was initiated using 3 mM MgSO4. The measurements for hydrolysis and transport activities of H+-ATPase were repeated three times, and mean values are given.
Construction of PeHA1-pro::GUS and transformation to tobacco
PeHA1-pro, the promoter of PeHA1, was cloned into the PMD18-T vector (Takara) and then transferred to a PBI121 vector. The promoter of PeHA1 was merged with the GUS gene and transformed into A. tumefaciens GV3101 (Shen et al., 2015a). PBI121-PeHA1-pro::GUS was finally transferred into WT tobacco using the leaf disc method (Shen et al., 2015a). We selected the putative transgenic plants on MS medium containing 200 mg l−1 kanamycin. Tobacco plantlets were incubated in a growth chamber at 25±1 °C with a relative humidity of 50–60%, a photoperiod of 16 h light/8 h dark, and a light intensity of 50 μmol m−2 s-1. WT and PeHA1-pro::GUS-transgenic tobacco (T1) seeds were allowed to germinate in MS medium. After culturing for 4 weeks, seedlings were treated with 0 or 200 mM NaCl for 24 h. Root, leaf, and stem tissues were sampled for GUS staining. Three individual plants for each treatment were used for GUS assays.
In situ localization of GUS
Root, leaf, and stem samples from PeHA1-pro::GUS transgenic tobacco and WT plants were soaked in X-Gluc solution {50 mM sodium phosphate buffer pH 7.0, 10 mM Na2EDTA, 0.5 mM K4[Fe(CN)6]·3H2O; 0.5 mM K3[Fe(CN)6], 0.1% Triton X-100, and 1 mg mL−1 X-Gluc} at 37 °C for 12 h (Shen et al., 2015a). The chlorophyll in the samples was removed with increasing concentrations of ethanol (70%→100%) at room temperature. The samples were fixed with FAA fixative (1:18:1 mix of formalin:alcohol:acetic acid), followed by gradient dehydration with increasing ethanol solutions (70%→85%→95%→100%→100%, 1−2 h for each ethanol concentration).
The protocols for subsequent infiltration were as follows: an equal volume mixture of dimethylbenzene and ethanol for 1 h→pure dimethylbenzene for 1 h→pure dimethylbenzene for 1 h. The samples were embedded with paraffin solution, and the wax blocks were trimmed to a regular trapezoid shape using a razor blade. Sections with a thickness of 25 μm were cut using an automatic vibration slicer (Leica VT1000 S, Leica Instruments, Nussloch, Germany). The sections were stuck on a glass slide and baked at 40 °C. Dewaxing was performed as follows (5–10 min for each chemical treatment): dimethylbenzene→1/2 dimethylbenzene+1/2 pure ethanol→100% ethanol→95% ethanol (rapid)→100% ethanol→1/2 dimethylbenzene+1/2 pure ethanol→dimethylbenzene. An appropriate amount (1−2 drops) of neutral gum was added to the slide, which was covered with a clean coverslip. The sections were incubated at 42 °C, dried overnight, and detected using an Olympus BX43 light microscope (Olympus Corporation, Tokyo, Japan). We used a stereomicroscope (LeicaM205 C, Leica, Wetzlar, Germany) to observe the samples and acquire images. Representative images are shown.
Protein extraction and fluorometric GUS assays
Based on Jefferson’s protocols (Jefferson, 1987), GUS activity in PeHA1-pro transgenic tobacco leaves (with main veins), stems, and roots was determined using 4-methyl-umbelliferyl glucuronide (Sigma-Aldrich) as a substrate for fluorescence assays. The protein concentration of the extract was determined using BSA as a standard protein in a microplate fluorescence spectrophotometer (M200, Tecan Group Ltd, Männedorf, Switzerland). The experimentally detected excitation wavelength was 365 nm, and the emission wavelength was 455 nm.
Statistical analysis
The data were subjected to ANOVA. Significant differences between means were determined by Duncan’s multiple range test. Unless otherwise stated, P<0.05 was considered significant.
Results
Salt-induced transcription of PeHA1 in P. euphratica
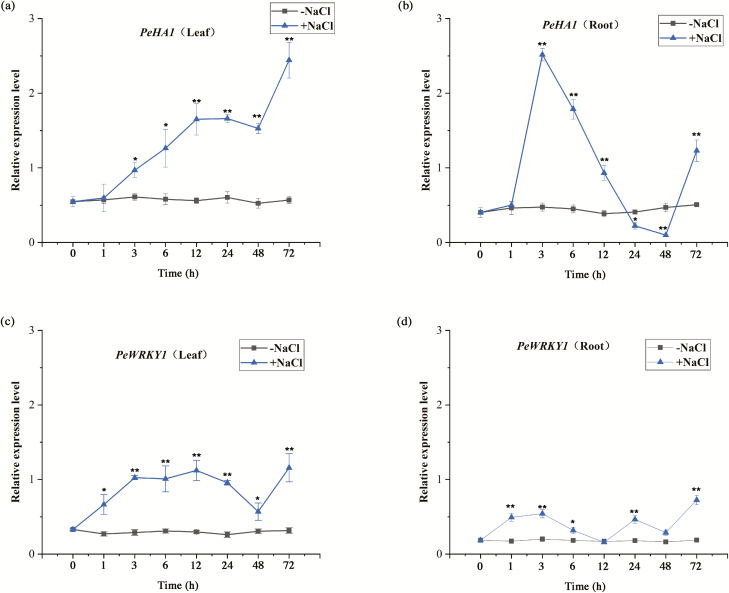
NaCl markedly increased the expression of HA1 in P. euphratica, though the pattern differed between roots and leaves (Fig. 1). In leaves, PeHA1 steadily increased after the onset of salt exposure, reaching a maximum level at 72 h, although a decline was observed at 24−48 h (Fig. 1a). In roots, an earlier and abrupt increase occurred after 3 h of salt treatment, followed by a significant decline at 24 h and 48 h, then another increase at 72 h (Fig. 1b).
Fig. 1.
Expression pattern of PeWRKY1 and PeHA1 in the roots and leaves of Populus euphratica treated with NaCl. P. euphratica seedlings were subjected to 200 mM NaCl for 72 h. (a) PeHA1 in leaves. (b) PeHA1 in roots. (c) PeWRKY1 in leaves. (d) PeWRKY1 in roots. Expression levels of PeWRKY1 and PeHA1 were normalized to the P. euphratica housekeeping gene PeACT7 as an internal reference. Primers designed to target PeWRKY1, PeHA1, and PeACT7 are shown in Supplementary Table S1. Each point corresponds to the mean of three independent replicates, and bars represent the SEM. *P<0.05, **P<0.01. (This figure is available in color at JXB online.)
PeHA1 promoter analysis
To determine the importance of the upstream regulatory region for salt-induced PeHA1 expression, a 2022 bp promoter fragment upstream of PeHA1 was sequenced and analyzed using the PLACE and Plant-CARE databases (Supplementary Fig. S1). The promoter sequence contained various cis-acting elements, including MRE (MYB-binding site involved in light responsiveness), MYC (cis-acting element responsive to drought and abscisic acid), P-box (gibberellin-responsive element), LTR (cis-acting element involved in low-temperature responsiveness), CGTCA-motif (cis-acting regulatory element involved in methyl jasmonate responsiveness), ARE (cis-acting regulatory element essential for anaerobic induction), GC-motif (enhancer-like element involved in anoxic-specific inducibility), MYB (gene element involved in response to drought and ABA signals), G-box (cis-acting regulatory element involved in light responsiveness), and ABRE (transcription factors involved in abscisic acid responsiveness). Interestingly, a cis-acting element W-box (T)TGAC(C), the binding site for the WRKY family of transcription factors, was present in the promoter region of PeHA1 (Supplementary Fig. S1).
NaCl activated the PeHA1 promoter in root and shoot tissues
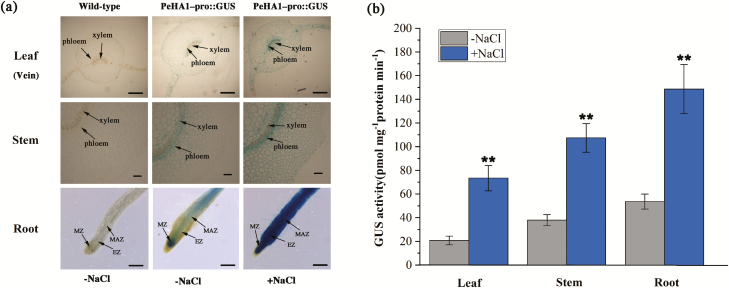
The PeHA1-pro::GUS fusion was constructed and transferred to tobacco leaves. GUS activity was observed in the root tips, leaf mesophyll and veins, and vascular tissues of the stem (Fig. 2a). NaCl treatment (200 mM, 24 h) significantly increased GUS activity in the root, leaf, and stem tissues (Fig. 2b). Therefore, the salt-enhanced expression of PeHA1 in P. euphratica was shown to be due to promoter activity.
Fig. 2.
Anatomical and histochemical analysis of GUS activity in PeHA1-pro::GUS-transgenic plants. Four-week-old seedlings of PeHA1-pro::GUS-transgenic plants were treated with 0 (−NaCl) or 200 mM NaCl (+NaCl) for 24 h. The wild-type (WT) controls were not treated with NaCl. Root, leaf, and stem samples were taken from control and salinized plants and used for anatomic analysis. (a) GUS staining. Scale bar=500 μm. (b) GUS activity was detectable in PeHA1-pro::GUS-transgenic plants regardless of the presence of salt stress, but was not detectable in WT plants. Each column corresponds to the mean of three independent replicates, and bars represent the SEM. **P<0.01. (This figure is available in color at JXB online.)
Salt-induced transcription of PeWRKY1 and DAP-seq
NaCl up-regulated PeWRKY1 transcription in both the roots and leaves (Fig. 1c, d). This result is similar to previous findings by Shen et al. (2015a), who found that the PeWRKY1 transcript level increased during a short-term salt treatment (12 h); however, PeWRKY1 expression returned to the control level after a long-term salt treatment (7 d). Notably, salt-elicited PeWRKY1 showed a trend similar to PeHA1, but with an earlier increase at 1 h in both roots and leaves (Fig. 1).
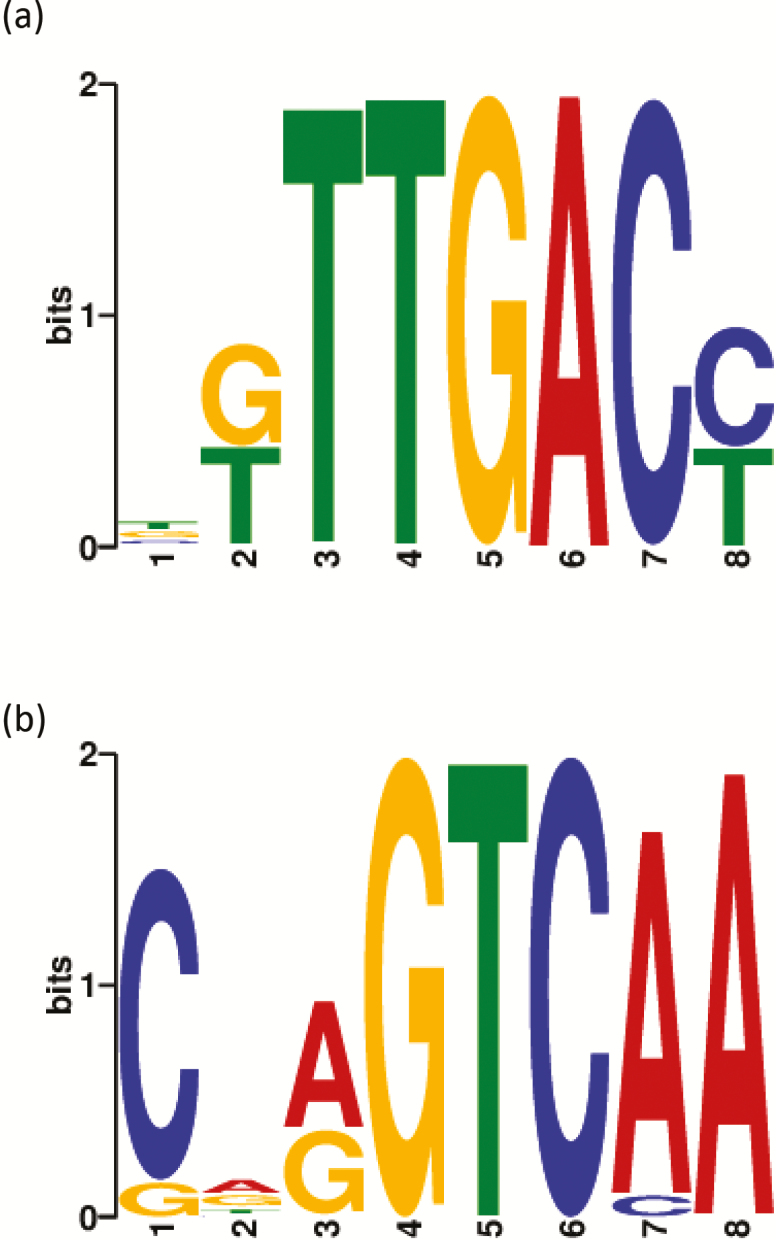
DAP-seq was performed to screen direct target genes of the transcription factor, PeWRKY1. The data showed that the top motifs for PeWRKY1-binding were ‘TTGAC’ and ‘GTCAA’, respectively (Fig. 3a, b). By screening the binding targets of the transcription factor, P. euphratica PM ATPase 4 (LOC105140044), which was named PeHA1 in our previous study (Wang et al., 2013) and in this work, was the target gene of PeWRKY1 protein (Supplementary Table S3).
Fig. 3.
Populus euphratica DNA motifs bound by PeWRKY1 protein in DNA affinity purification sequencing (DAP-seq). The two top logos for PeWRKY1 binding are shown in (a) and (b). P. euphratica leaves were sampled from each of three individual plants and subjected to DAP-seq. E-value (the statistical significance of the motif) 1.9e-108 or 1.9 × 10–108 (a). E value 5.2e-021 or 5.2 × 10–21 (b). The PeWRKY1 DAP-seq peaks located upstream of the transcription start site (TSS) of the PeHA1 gene are shown in Supplementary Table S3. (This figure is available in color at JXB online.)
PeWRKY1 binds to the W-box of the PeHA1 promoter
To determine whether PeWRKY1 interacts with PeHA1, Y1H assay, EMSA, LRA, and VIGS were performed to confirm the binding of PeWRKY1 to the W-box in the promoter region of PeHA1.
Y1H assay
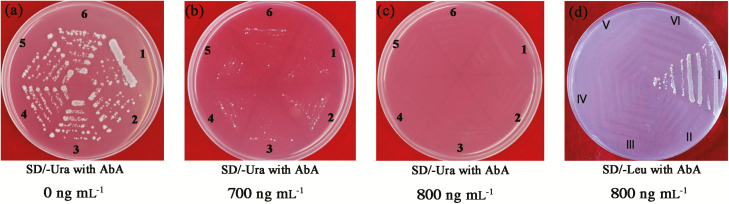
A 1266 bp promoter sequence of PeHA1 was fused to the yeast vector (pAbAi) with the reporter gene AUR1-C (Shen et al., 2015a). pAbAi-PeHA1pro grew on SD/-Ura medium containing 0−700 ng ml−1 AbA, but was inhibited at 800 ng ml−1 AbA (Fig. 4a–c). However, the Y1H yeast strain grew on SD/-Leu medium (800 ng ml−1 AbA) with the plasmid carrying PeWRKY1 (Fig. 4d). The Y1H test demonstrated that PeWRKY1 can interact with PeHA1-pro.
Fig. 4.
Yeast one-hybrid analysis of PeWRKY1 binding to the PeHA1 promoter. The 1266 bp promoter of PeHA1 was cloned and fused into the pAbAi vector AUR1-C gene (with Aureobasidin A resistance). The synthetic W-box mutant promoter sequence (TTGACC mutated into TTGAAC) of PeHA1 was used as a control. (a–c) pAbAi (PeHA1pro)-Y1H (1, 2, 3, and 4) and control (5 and 6) strains were grown on SD/-Ura medium supplemented with 0, 700, or 800 ng ml−1 AbA. Yeast strains 1, 2, 3, 4, 5, and 6 could not grow on SD medium lacking Ura in the presence of 800 ng ml−1 AbA, which was used as the screening concentration in the PeWRKY1–W-box interaction experiment. (d) The pAbAi (PeHA1pro)-Y1H strain was transferred to the pGADT7 plasmid with or without PeWRKY1, and the PeWRKY1–W-box interaction was detected by growth on SD lacking Leu (800 ng ml–1 AbA). I, pAbAi (W-box)-Y1H-pGADT7 (PeWRKY1); II, Y1H-pGADT7; III, pAbAi-Y1H-pGADT7; IV, pAbAi (W-box)-Y1H-pGADT7; V, pAbAi (mutated W-box)-Y1H-pGADT7; VI, pAbAi-Y1H-pGADT7 (PeWRKY1). (This figure is available in color at JXB online.)
W-box deletion confirmed that PeWRKY1 bound only the W-box cis-acting element. Growth of the pAbAi-W-box mutant strain was severely inhibited by AbA in the presence and absence of PeWRKY1 plasmids (Fig. 4d). Therefore, PeWRKY1 could bind the W-box in the upstream regulatory region of the PeHA1 promoter.
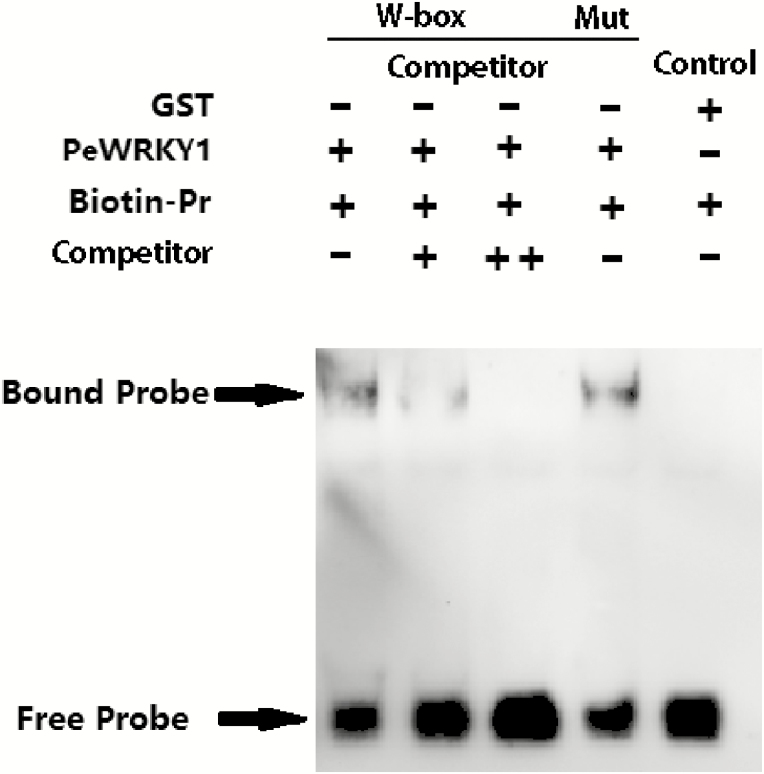
EMSA
The assay was performed with biotin-labeled W-box probes and unlabeled competitive probes. Biotin labeling showed PeWRKY1 protein binding to the W-box motif in the PeHA1 promoter, whereas the negative control, GST, could not bind to biotin-labeled W-box probes (Fig. 5, lanes 1 and 5). The increasing fraction of competitive probes interfered with the binding of PeWRKY1 to biotin-labeled W-box (Fig. 5, lanes 1−3). The binding specificity of the W-box for PeWRKY1 was confirmed with a mutated W-box. The W-box mutation probes were obtained with a double-stranded nucleotide mutation and not labeled with biotin. However, the presence of W-box mutation probes did not inhibit the binding of PeWRKY1 to biotin-labeled W-box (Fig. 5, lane 4).
Fig. 5.
An EMSA validated the PeWRKY1 interaction with the W-box in the PeHA1 promoter. The purified PeWRKY1–GST protein obtained by prokaryotic expression was used for the in vitro assay, and GST protein was used as a negative control. The purified proteins were combined with biotinylated W-box-containing dsDNA probe (1×), and unlabeled probes (Competitor) were subjected to cold competition experiments. The biotin-unlabeled mutant probes (Mut, TTGACC mutated into TTAGCC) were used to confirm the binding specificity of the W-box to PeWRKY1. ‘+’ and ‘–’ indicate the presence or absence, respectively, of proteins and probes in the loading mixture. The cold probe concentrations are 5× and 10×, and that of the Mut probe is 5×. The concentration of the polyacrylamide gel was 4%.
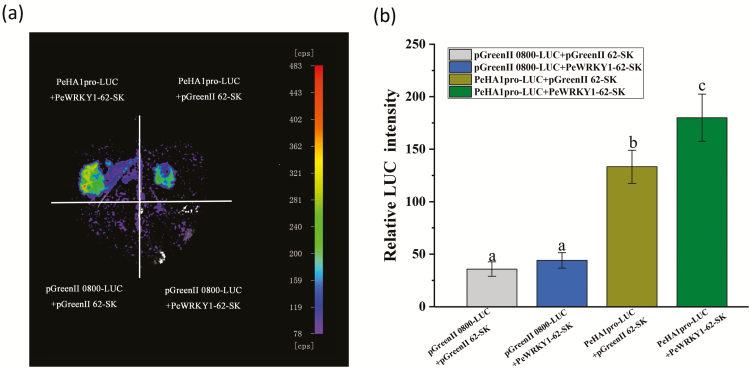
LRA
Tobacco leaves co-transformed with A. tumefaciens vector controls, pGreenII 0800-LUC and pGreenII 62-SK, showed very low luciferase luminescence (Fig. 6a, b). Co-transformation of pGreenII 0800-LUC and PeWRKY1-62-SK exhibited a low luminescence, similar to vector controls (Fig. 6a, b). Luciferase luminescence was observed when PeHA1pro-LUC was transformed into tobacco leaves; and the luminescence was significantly enhanced by the co-transformation of PeWRKY1 (Fig. 6a, b). The LRA confirmed that PeWRKY1 activated the PeHA1 promoter in N. benthamiana leaves.
Fig. 6.
The luciferase reporter assay affirmed that PeWRKY1 activated the PeHA1 promoter in Nicotiana benthamiana leaves. (a) Representative luciferase luminescence image of N. benthamiana leaves co-infiltrated with the agrobacterial strains containing PeHA1pro-Luc and PeWRKY1-62-SK. Tobacco leaves injected with empty vector controls, pGreenII 0800-LUC and pGreenII 62-SK, were used as a negative control. (b) The luminescence intensity in Agrobacterium-injected N. benthamiana leaves. Each column corresponds to the mean of three independent replicates, and bars represent the SEM. Columns labeled with different letters denote a significant difference (P<0.05). (This figure is available in color at JXB online.)
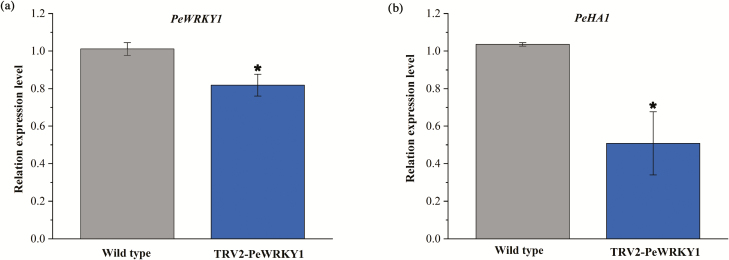
VIGS
TRV-based factor pTRV2 was used for VIGS in P. euphratica leaves, as described previously by Shen et al. (2015a, b). The expression of PeWRKY1 in leaves decreased after infiltration of Agrobacterium carrying TRV2-PeWRKY1 (Fig. 7a). Notably, the quantitative RT-PCR assay showed that the expression of PeHA1 in salinized leaves significantly decreased after 45 d of Agrobacterium infiltration (Fig. 7b). This indicates that PeWRKY1 silencing resulted in the decline of PeHA1 under saline conditions.
Fig. 7.
Expression of PeWRKY1 and PeHA1 in leaves of PeWRKY1-silenced Populus euphratica. (a) PeWRKY1 expression. P. euphratica leaves were infiltrated with Agrobacterium carrying TRV2-PeWRKY1, whereas the wild-type controls were transiently injected with a TRV2 empty vector. (b) PeHA1 expression in PeWRKY1-silenced P. euphratica seedlings. After 45 d of TRV infection, seedlings were subjected to 200 mM NaCl for 6 h. Expression levels of PeWRKY1 and PeHA1 were examined by quantitative RT-PCR. The P. euphratica housekeeping gene PeACT7 was used as an internal reference. Primers designed to target PeWRKY1, PeHA1, and PeACT7 are shown in Supplementary Table S1. Each column corresponds to the mean of three independent replicates, and bars represent the SEM. *P<0.05. (This figure is available in color at JXB online.)
Overexpression of PeWRKY1 enhanced salt tolerance in tobacco
In hydroponic culture, PeWRKY1-transgenic lines L-9 and L-12 exhibited a greater ability to tolerate salt stress than the WT and VC (Supplementary Fig. S3a). Leaf fresh weight was less suppressed by NaCl and the increase in MDA was lower in transgenic lines than in salt-stressed WT and VC (Supplementary Fig. S3b, c). The salt-elicited oxidative damage and inhibited growth in WT and VC plants were due to excessive accumulation of Na+ ions. Na+ concentrations were 36−52% lower in the transgenic lines than in WT and VC plants under salt treatment (Supplementary Fig. S4a). The lower Na+ build up was due to the higher Na+ efflux in salinized roots of the transgenic lines (Supplementary Fig. S4b).
PeWRKY1 enhanced NtHA transcripts and H+-ATPase activity
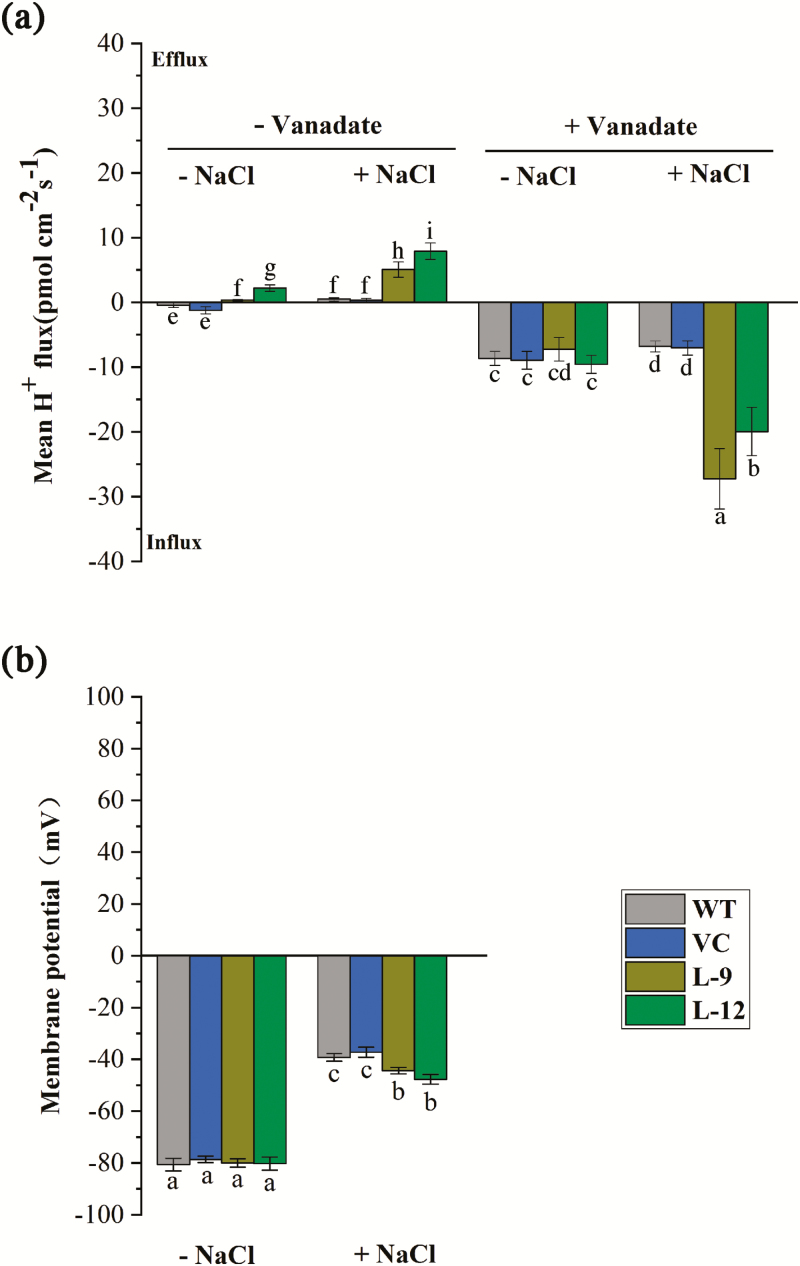
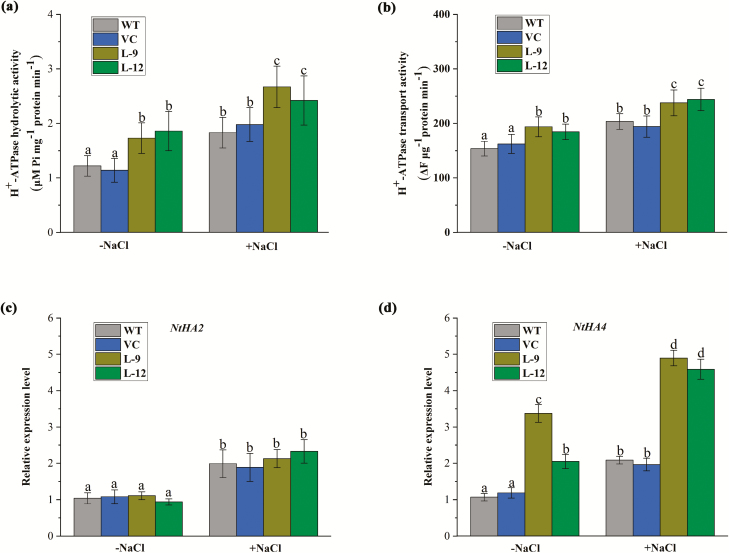
NaCl increased H+ efflux in the root tips, and the effect was more pronounced in the transgenic lines (Fig. 8a). However, the salt-elicited H+ efflux shifted towards a net influx by vanadate, a specific inhibitor of H+ pumps in the PM (Fig. 8a). Thus, PeWRKY1 promoted proton pumping activity under salinity stress. The membrane potential of root cells was also measured by NMT microelectrodes. The two transgenic lines exhibited a more negative membrane potential than the WT and VC plants, though the membrane hyperpolarization decreased with NaCl treatment (200 mM, 24 h; Fig. 8b). The PM vesicles were isolated from tobacco roots and used for the H+-ATPase activity assay. The hydrolytic and transport activities of H+-ATPase were higher in transgenic lines than in the WT and VC, regardless of whether they were under control or NaCl stress conditions (Fig. 9a, b).
Fig. 8.
H+ fluxes and membrane potential in roots of wild-type (WT) tobacco and PeWRKY1-transgenic plants under salt stress. (a) H+ fluxes at the root tip. WT, vector control (VC), and transgenic lines (L-9 and L-12) were subjected to 0 (–NaCl) or 200 mM NaCl (+NaCl) for 24 h. The roots were then exposed to sodium orthovanadate (500 μM) for 30 min. Steady-state H+ fluxes were continuously recorded for 10 min at the apical zones (200 μm from the root tip) in the absence or presence of the inhibitor. (b) Membrane potential in control and salinized roots was continuously recorded for 10 min at the apical region (200 µm from the tip). Each column is the mean of 5–6 individual plants, and error bars represent the SE. Columns labeled with different letters denote a significant difference (P<0.05). (This figure is available in color at JXB online.)
Fig. 9.
H+-ATPase activity and gene expression in wild-type (WT) tobacco and PeWRKY1-transgenic plants under salt stress. WT, vector control (VC), and transgenic lines (L-9 and L-12) were subjected to 0 (–NaCl) or 200 mM NaCl (+NaCl) for 24 h. Plasma membrane vesicles were isolated from the control and salinized plants and used to measure hydrolytic activity and H+ transport activity. (a) ATPase hydrolytic activity measured according to the Pi released from ATP. (b) Proton transport activity of ATPase measured by fluorescent quenching of acridine orange at 495 nm. (c and d) Expression of NtHA2 and NtHA4. Expression levels of NtHA2 (c) and NtHA4 (d) were examined by quantitative RT-PCR. The tobacco housekeeping gene, NtEF1α, was used as an internal reference. Primers designed to target NtHA2, NtHA4, and NtEF1α are shown in Supplementary Table S1. Each column corresponds to the mean of three independent replicates, and bars represent the SEM. Columns labeled with different letters denote a significant difference (P<0.05). (This figure is available in color at JXB online.)
Quantitative RT-PCR showed that NaCl up-regulated the expression of PM H+-ATPase-encoding genes NtHA2 and NtHA4 (Fig. 9c, d). Notably, the transcription of NtHA4 was significantly higher in the transgenic lines than in the WT and VC plants in the absence or presence of salt stress (Fig. 9d). We found a W-box in the upstream regulatory region of NtHA4 (Supplementary Fig. S5) but not in the promoter fragment of NtHA2 (Supplementary Fig. S6). Therefore, PeWRKY1 may interact with the W-box and accelerate the transcription of NtHA4.
Discussion
NaCl-elicited WRKY1 mediates changes in HA1 expression in P. euphratica
Salt-tolerant P. euphratica retain high proton pumping activity to promote Na+ extrusion under salt stress (Sun et al., 2009a, b, 2010a, b, 2012). Maintaining high expression of ATPase genes is crucial for P. euphratica to withstand long-term salt exposure, although regulation of the PM ATPases is primarily post-transcriptional in Arabidopsis (Palmgren et al., 1991; Svennelid et al., 1999; Camoni et al., 2000; Yang et al., 2010). Janz et al. (2010) showed that P. euphratica exhibits permanent activation of numerous genes responsible for salt exclusion and ion compartmentalization. We have previously shown that control P. euphratica plants exhibit a higher transcript abundance of genes related to Na+/H+ antiport (Na+/H+ antiporters, H+ pumps) compared with the salt-sensitive poplar, and the expression of these H+ pump genes did not decrease or was even up-regulated during salt treatment (Ding et al., 2010). Up-regulating the expression of the PM H+-ATPase gene increased salt tolerance in Arabidopsis (Gévaudant et al., 2007; Wang et al., 2013) and in A. littoralis (Olfatmiri et al., 2014). Here, we showed that NaCl increased the expression of PeHA1 in P. euphratica roots and leaves (Fig. 1). Anatomical and histochemical GUS analyses showed that the PeHA1 promoter became active in the roots and shoots when the plants were exposed to NaCl (Fig. 2). Therefore, the salt-enhanced expression of PeHA1 in P. euphratica was apparently due to the promoter activity. Notably, PeHA1 contained a W-box sequence in the promoter (Supplementary Fig. S1), which can be recognized by PeWRKY1 (Rushton et al., 1996; Eulgem et al., 2000; Rushton et al., 2010). NaCl increased expression of PeWRKY1 in P. euphratica roots and leaves (Fig. 1), which is in agreement with our previous findings in this salt-resistant poplar (Shen et al., 2015a). Similarly, 23 of the 107 WRKY genes can be altered by salt stress in the P. euphratica genome (Ma et al., 2015). PeHA1 induced by salt stress follows a similar trend to PeWRKY1 in both the roots and leaves (Fig. 1).
DAP-seq revealed PeHA1 as the target gene of the PeWRKY1 protein (Fig. 3; Supplementary Table S3). Furthermore, we confirmed that PeWRKY1 bound the W-box of the PeHA1 promoter by Y1H assay (Fig. 4), EMSA (Fig. 5), and LRA (Fig. 6). PeHA1 was down-regulated when PeWRKY1 was silenced in salinized P. euphratica leaves (Fig. 7). Overall, our data revealed that PeWRKY1 activated PeHA1 expression by binding to the cis-acting W-box of the PeHA1 promoter in salt-stressed P. euphratica. In rice, WRKY13 physically binds to the promoter region of SNAC11, which harbors W or W-like box-type cis-regulatory elements for the binding of WRKY transcription factors (Qiu et al., 2009; Tao et al., 2009).
PeWRKY1 interacts with PeHA1, contributing to increased H+ pumping activity and Na+ exclusion
PeWRKY1 overexpression enhanced salt tolerance in transgenic plants due to the lower amount of accumulated Na+ (Supplementary Figs. S3, S4). Similarly, Shen et al. (2015a) showed that Na+ efflux was significantly enhanced in transgenic plants overexpressing PeWRKY1. Na+ exclusion is critical for salt adaption. Excessive accumulation of Na+ increased lipid peroxidation, as indicated by MDA content (Supplementary Figs. S3, S4; Wang et al., 2007). We have shown that active Na+ extrusion is promoted by the H+ pumps in the PM (Wang et al., 2013). PeWRKY1 overexpression enhanced the activity of PM H+-ATPases in both in vivo assays with NMT microelectrodes and in vitro assays with isolated PM vesicles (Figs 8, 9).
NaCl caused a pronounced H+ efflux across the PM in the transgenic lines as the result of activated PM H+-ATPase, which also caused a negative membrane potential in salinized roots (Fig. 8). In vitro H+-ATPase assays were consistent with in vivo observations (Fig. 9). The activated proton pumps maintained the membrane potential under salt stress and provided the driving force for Na+ extrusion, thereby avoiding excessive accumulation of Na+ (Chen et al., 2007; Sun et al., 2009b, 2010a). The high H+-pumping activities resulted, at least in part, from increased HA transcripts in transgenic tobacco. Our previous studies showed that PeHA1 overexpression increases the hydrolysis and transport activities of proton pumps in Arabidopsis response to salt stress (Wang et al., 2013). Moreover, PeWRKY1 enhanced Na+/H+ antiport in transgenic plants, which is presumably related to the H+ pumps (Shen et al., 2015a). NtHA2 and NtHA4 were up-regulated by NaCl in tobacco plants, with NtHA4 significantly more up-regulated in transgenic plants than in the WT and VC, under both control and saline conditions (Fig. 9). It is likely that PeWRKY1 bound to the W-box in the promoter of NtHA4, enhancing its expression. Consequently, PeWRKY1 improved the activity of PM H+-ATPase, resulting in an increased capacity to tolerate salinity stress. In rice, WRKY13 binds to W-like boxes to regulate gene expression during abiotic and biotic stress (Xiao et al., 2013). In ChIP-quantitative PCR analyses, Hu et al. (2013) showed that WRKY8 interacts with one W-box element in the promoter of RD29A, directly regulating RD29A transcription under salt stress.
Taken together, the results indicate that PeWRKY1 activated PeHA1 transcription by binding to the promoter region, conferring H+ pumping and Na+ homeostasis control and salt tolerance. Further investigation is needed to determine whether the PeHA1-encoded protein was post-transcriptionally regulated in salinized P. euphratica.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Promoter sequence analysis of Populus euphratica PeHA1.
Fig. S2. Semi-quantitative RT-PCR analysis of PeWRKY1 in wild-type (WT) tobacco and transgenic lines L1–L15.
Fig. S3. Phenotype tests of wild-type (WT) tobacco and PeWRKY1-transgenic plants.
Fig. S4. Na+ concentrations and fluxes in roots of wild-type (WT) tobacco and PeWRKY1-transgenic plants.
Fig. S5. Promoter sequence analysis of tobacco NtHA4.
Fig. S6. Promoter sequence analysis of tobacco NtHA2.
Table S1. Primer sets used for quantitative real-time PCR.
Table S2. Primers used for cloning the promoters of Populus euphratica PeHA1 and Nicotiana tabacum L. NtHA2 and NtHA4.
Table S3. PeWRKY1 DAP-seq peaks located upstream of the transcription start site (TSS) of the PeHA1 gene.
Data availability
Physiological data were collected from at least three replicates from wild-type genotype and transgenic plants. Dryad Digital Repository. https://doi.org/10.5061/dryad.83ng085; Yao et al. 2020.
Supplementary Material
Acknowledgements
The research was supported jointly by the National Natural Science Foundation of China (grant nos 31770643 and 31570587), Beijing Natural Science Foundation (grant nos 6182030 and 6172024), the Fundamental Research Funds for the Central Universities (grant no. 2019ZY25), the Program of Introducing Talents of Discipline to Universities (111 Project, grant no. B13007), and the Beijing Advanced Innovation Center for Tree Breeding by Molecular Design (Beijing Forestry University). Prof. Dr Yule Liu (School of Life Sciences, Tsinghua University) is sincerely acknowledged for providing TRV vectors.
Glossary
Abbreviations:
- AbA
Aureobasidin A
- DAP-seq
DNA affinity purification sequencing
- EMSA
electrophoretic mobility shift assay
- GUS
β-glucuronidase
- LRA
luciferase reporter assay
- MDA
malondialdehyde
- NMT
non-invasive microtest
- PM
plasma membrane
- TCA
trichloroacetic acid
- VC
vector control
- VIGS
virus-induced gene silencing
- WT
wild type
- Y1H
yeast one-hybrid.
Author contributions
JY, ZS, and SC conceived and designed the experiments; JY, Y-LZ, XW, JW, and GS performed the experiments; JY, JW, RZ, Y-HZ, HZ, CD, JL, SH, YZ, Y-NZ, NZ, and SD analyzed the experimental data; JY, RZ, and SL wrote the article; and SC revised the article.
Conflict of interest
All authors have no conflict of interest to declare.
References
- Bartlett A, O’Malley R, Huang S, Galli M, Nery J, Gallavotti A, Ecker J. 2017. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nature Protocols 12, 1659−1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camoni L, Iori V, Marra M, Aducci P. 2000. Phosphorylation-dependent interaction between plant plasma membrane H+-ATPase and 14-3-3 proteins. Journal of Biological Chemistry 275, 9919–9923. [DOI] [PubMed] [Google Scholar]
- Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X. 2010. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biology 10, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. 2008. Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiology 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D. 2012. The role of WRKY transcription factors in plant abiotic stresses. Biochimica et Biophysica Acta 1819, 120–128. [DOI] [PubMed] [Google Scholar]
- Chen S, Li J, Wang S, Hüttermann A, Altman A. 2001. Salt, nutrient uptake and transport, and ABA of Populus euphratica; a hybrid in response to increasing soil NaCl. Trees 15, 186−194. [Google Scholar]
- Chen S, Polle A. 2010. Salinity tolerance of Populus. Plant Biology 12, 317–333. [DOI] [PubMed] [Google Scholar]
- Chen SL, Hawighorst P, Sun J, Polle A. 2014. Salt tolerance in Populus: significance of stress signaling networks, mycorrhization, and soil amendments for cellular and whole-plant nutrition. Environmental and Experimental Botany 107, 113−124. [Google Scholar]
- Chen ZH, Pottosin IP, Cuin AT, et al. . 2007. Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiology 145, 1714−1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Hou P, Shen X, et al. . 2010. Salt-induced expression of genes related to Na+/K+ and ROS homeostasis in leaves of salt-resistant and salt-sensitive poplar species. Plant Molecular Biology 73, 251–269. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. 2000. The WRKY superfamily of plant transcription factors. Trends in Plant Science 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Gévaudant F, Duby G, von Stedingk E, Zhao R, Morsomme P, Boutry M. 2007. Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiology 144, 1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM. 2013. Sodium (Na+) homeostasis and salt tolerance of plants. Environmental and Experimental Botany 92, 19−31. [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YR, Chen LG, Wang HP, Zhang LP, Wang F, Yu DQ. 2013. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. The Plant Journal 74, 730−745. [DOI] [PubMed] [Google Scholar]
- Ishihama N, Yamada R, Yoshioka M, Katou S, Yoshioka H. 2011. Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. The Plant Cell 23, 1153–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz D, Behnke K, Schnitzler JP, Kanawati B, Schmitt-Kopplin P, Polle A. 2010. Pathway analysis of the transcriptome and metabolome of salt sensitive and tolerant poplar species reveals evolutionary adaption of stress tolerance mechanisms. BMC Plant Biology 10, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Lu J, Xu J, Duan B, He X, Liu J. 2015. Genome-wide identification of WRKY genes in the desert poplar Populus euphratica and adaptive evolution of the genes in response to salt stress. Evolutionary Bioinformatics Online 11, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Wang J, Zhou G, et al. . 2013. Genomic insights into salt adaptation in a desert poplar. Nature Communications 4, 2797. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olfatmiri H, Alemzadeh A, Zakipour Z. 2014. Up-regulation of plasma membrane H+-ATPase under salt stress may enable Aeluropus littoralis to cope with stress. Molecular Biology Research Communications 3, 67–75. [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166, 1598. [DOI] [PubMed] [Google Scholar]
- Palmgren MG. 1990. An H+-ATPase assay: proton pumping and ATPase activity determined simultaneously in the same sample. Plant Physiology 94, 882–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C. 1991. Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. Journal of Biological Chemistry 266, 20470–20475. [PubMed] [Google Scholar]
- Pitman M, Läuchli A. 2002. Global impact of salinity and agricultural ecosystems. Salinity: environment–plants–molecules. Dordrecht: Springer, 3−20 [Google Scholar]
- Polle A, Chen S. 2015. On the salty side of life: molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant, Cell & Environment 38, 1794–1816. [DOI] [PubMed] [Google Scholar]
- Qin YX, Tian YC, Han L, Yang XC. 2013. Constitutive expression of a salinity-induced wheat WRKY transcription factor enhances salinity and ionic stress tolerance in transgenic Arabidopsis thaliana. Biochemical and Biophysical Research Communications 441, 476−481. [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Cheng H, Li X, Wang S. 2009. Exploring transcriptional signalling mediated by OsWRKY13, a potential regulator of multiple physiological processes in rice. BMC Plant Biology 9, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. 2010. WRKY transcription factors. Trends in Plant Science 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. 1996. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. The EMBO Journal 15, 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Sa G, Yao J, Deng C, et al. . 2019. Amelioration of nitrate uptake under salt stress by ectomycorrhiza with and without a Hartig net. New Phytologist 222, 1951–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. . 2002. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal 31, 279–292. [DOI] [PubMed] [Google Scholar]
- Shen Z, Yao J, Sun J, et al. . 2015. a Populus euphratica HSF binds the promoter of WRKY1 to enhance salt tolerance. Plant Science 235, 89–100. [DOI] [PubMed] [Google Scholar]
- Shen ZD, Ding M, Sun J, et al. . 2013. Overexpression of PeHSF, mediates leaf ROS homeostasis in transgenic tobacco lines grown under salt stress conditions. Plant Cell, Tissue and Organ Culture 115, 299−308. [Google Scholar]
- Shen ZD, Sun J, Yao J, et al. . 2015. b High rates of virus-induced gene silencing by tobacco rattle virus in Populus. Tree Physiology 35, 1016−1029. [DOI] [PubMed] [Google Scholar]
- Song X, Duanmu H, Yu Y, et al. . 2017. GsJ11, identified by genome-wide analysis, facilitates alkaline tolerance in transgenic plants. Plant Cell, Tissue and Organ Culture 129, 411−430. [Google Scholar]
- Sun J, Chen S, Dai S, et al. . 2009. a NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiology 149, 1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Dai SX, Wang RG, et al. . 2009. b Calcium mediates root K+/Na+ homeostasis in poplar species differing in salt tolerance. Tree Physiology 29, 1175−1186. [DOI] [PubMed] [Google Scholar]
- Sun J, Li LS, Liu MQ, et al. . 2010. b Hydrogen peroxide and nitric oxide mediate K+/Na+ homeostasis and antioxidant defense in NaCl-stressed callus cells of two contrasting poplars. Plant Cell, Tissue and Organ Culture 103, 205−215. [Google Scholar]
- Sun J, Wang MJ, Ding MQ, et al. . 2010. a H2O2 and cytosolic Ca2+ signals triggered by the PM H+-coupled transport system mediate K+/Na+ homeostasis in NaCl-stressed Populus euphratica cells. Plant, Cell & Environment 33, 943–958. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhang X, Deng S, et al. . 2012. Extracellular ATP signaling is mediated by H2O2 and cytosolic Ca2+ in the salt response of Populus euphratica cells. PLoS One 7, e53136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M. 1999. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. The Plant Cell 11, 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S. 2009. A pair of allelic WRKY genes play opposite roles in rice–bacteria interactions. Plant Physiology 151, 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang Y, Sun J, et al. . 2013. Overexpression of PeHA1 enhances hydrogen peroxide signaling in salt-stressed Arabidopsis. Plant Physiology and Biochemistry 71, 37–48. [DOI] [PubMed] [Google Scholar]
- Wang RG, Chen SL, Deng L, Fritz E, Hüttermann A, Polle A. 2007. Leaf photosynthesis, fluorescence response to salinity and the relevance to chloroplast salt compartmentation and anti-oxidative stress in two poplars. Trees - Structure and Function 21, 581−591. [Google Scholar]
- Wang SJ, Wang JY, Yao WJ, Zhou BR, Li RH, Jiang TB. 2014. Expression patterns of WRKY genes in di-haploid Populus simonii × P. nigra in response to salinity stress revealed by quantitative real-time PCR and RNA sequencing. Plant Cell Reports 33, 1687−1696. [DOI] [PubMed] [Google Scholar]
- Xiao J, Cheng H, Li X, Xiao J, Xu C, Wang S. 2013. Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiology 163, 1868–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YQ, Qin YX, Xie CG, et al. . 2010. The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. The Plant Cell 22, 1313−1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Shen Z, Zhang Y, et al. . 2020. Data from: Populus euphratica WRKY1 binds the promoter of H+-ATPase gene to enhance gene expression and salt tolerance. Dryad Digital Repository. 10.5061/dryad.83ng085 [DOI] [PMC free article] [PubMed]
- Zhang Y, Liu T, Meyer CA, et al. . 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YN, Wang Y, Sa G, et al. . 2017. Populus euphratica J3 mediates root K+/Na+, homeostasis by activating plasma membrane H+-ATPase in transgenic Arabidopsis under NaCl salinity. Plant Cell, Tissue and Organ Culture 131, 75–88. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Physiological data were collected from at least three replicates from wild-type genotype and transgenic plants. Dryad Digital Repository. https://doi.org/10.5061/dryad.83ng085; Yao et al. 2020.