A gain-of-function rice mutant Photoperiod-thermo-sensitive dwarfism 1 is caused by a mutated gene, Ptd1, that encodes a nsLTP variant with an altered protein conformation due to disruption of the conserved eight-cysteine-motif.
Keywords: Disulfide bond, eight-cysteine motif, non-specific lipid transfer proteins (nsLTP), Photoperiod-thermo-sensitive dwarfism 1, plant height, rice
Abstract
Plant height is an important trait for architecture patterning and crop yield improvement. Although the pathways involving gibberellins and brassinosteroids have been well studied, there are still many gaps in our knowledge of the networks that control plant height. In this study, we determined that a dominant photoperiod- and thermo-sensitive dwarf mutant is caused by the active role of a mutated gene Photoperiod-thermo-sensitive dwarfism 1 (Ptd1), the wild-type of which encodes a non-specific lipid transfer protein (nsLTP). Ptd1 plants showed severe dwarfism under long-day and low-temperature conditions, but grew almost normal under short-day and high-temperature conditions. These phenotypic variations were associated with Ptd1 mRNA levels and accumulation of the corresponding protein. Furthermore, we found that the growth inhibition in Ptd1 may result from the particular protein conformation of Ptd1 due to loss of two disulfide bonds in the eight-cysteine motif (8-CM) that is conserved among nsLTPs. These results contribute to our understanding of the novel function of disulfide bonds in the 8-CM, and provide a potential new strategy for regulation of cell development and plant height by modifying the amino acid residues involved in protein conformation patterning.
Introduction
Plant height is an important trait for crop breeding. The ‘Green Revolution’ that was launched in late 1950s was characterized by breeding and large-scale cultivation of semi-dwarf crop varieties, and has achieved dramatic increases in grain yields, especially in wheat and rice (Evenson and Gollin, 2003). However, the widespread use of dwarfing germplasm of narrow genetic range combined with overuse of pesticides and fertilizers has led to serious environmental problems (Pimentel, 1996). This has triggered the examination of the genetic and molecular mechanisms that underlie height regulation with a view to constructing an ‘ideal’ plant architecture. It is known that most of the identified plant-height mutants are related to the metabolism or signaling of the phytohormones gibberellin (GA) and brassinosteroids (BRs), such as semi-dwarf1 (sd1) (Sasaki et al., 2002), gibberellins-insensitive dwarf1 (gid1) (Ueguchi-Tanaka et al., 2005), BR-deficient dwarf1 (brd1) (Mori et al., 2002), and brassinosteroid-insensitive1 (bri1) (Clouse et al., 1996). Although GA- and BR-related mutants have been studied extensively, an integrated picture of the molecular mechanisms responsible for the regulation of plant height has yet to be determined. An increasing number of novel plant-height mutants are being identified and characterized, and pathways other than those of GA and BR are being discovered. For instance, the carotenoid-derived phytohormone strigolactone has become a focus of research in plant architecture patterning (Zhou et al., 2013).
Non-specific lipid transfer proteins (nsLTPs) are a family of small and basic proteins widely present in land plants. The nsLTPs carry a highly conserved eight-cysteine motif (8-CM) with a common backbone of formula C-Xn-C-Xn-CC-Xn-CXC-Xn-C-Xn-C forming four disulfide bonds, which is essential for the conformation of a hydrophobic cavity. This cavity determines the binding of hydrophobic molecules such as fatty acids, phospholipids, glycolipids, and fatty acyl-CoA (Shinichiro and Mitsuhiro, 1986; Nichols, 1987; Tsuboi et al., 1992; Kader, 1996; José-Estanyol et al., 2004; Liu et al., 2015a; Salminen et al., 2016), and it plays roles in significant biological processes including disease resistance (Maldonado et al., 2002), formation of epidermal cutin and wax (DeBono et al., 2009; Lee et al., 2009), regulation of cell-wall loosening and extension (Nieuwland et al., 2005; Ambrose et al., 2013), anther development (Zhang et al., 2010), pollen development and germination (Chae et al., 2009), and stress resistance (Jung et al., 2005; Pitzschke et al., 2014).
We have previously identified a rice dominant dwarf mutant, Photoperiod-sensitive dwarf 1 (Psd1), which displays severe dwarfism when growing in long-days (LDs), but the height is partially recovered under short-day (SD) conditions (Li et al., 2014). The number of stem internodes of Psd1 is the same as the wild-type (WT), but the lengths of the elongated internodes are substantially shorter, due to a decrease in cell number and much shorter cell lengths of the internodes. The elongation of the seedling leaf-sheath of Psd1 can respond to a certain extent to exogenous bioactive GA. The WT gene of Psd1 encodes a nsLTP, while a single-nucleotide substitution and a single-nucleotide deletion in the near 3´-end coding region of Psd1 lead to a frame-shift, which results in a variant that lacks the last two Cys residues in the 8-CM motif and harbors a new 18-amino acid (aa) C-terminus. However, it is unclear how Psd1 exerts a dominant effect for growth inhibition, and what mechanisms underlie the photoperiod sensitivity of plant height.
Here, we have further characterized Psd1 and found that the dwarfism phenotype was also sensitive to temperature conditions, and thus we have renamed this mutant Photoperiod-thermo-sensitive dwarfism 1 (Ptd1). Our results indicate that the patterns of mRNA expression and protein accumulation account for the plant height phenotypes in response to different photoperiod and temperature conditions. We have also determined that the biochemical role of Ptd1 results from a specific protein conformation that is due to the loss of the last two disulfide bonds of the 8-CM.
Materials and methods
Plant materials
Ptd1 is a dominant dwarf mutant of rice (Oryza sativa) arising from a somaclonal variation of the japonica cultivar Zhonghua 11 (ZH11) (Li et al., 2014). The WT (PTD1/PTD1) and mutant homozygote (Ptd1/Ptd1) were selected from the segregants of a heterozygous mutant line (Ptd1/PTD1) by PCR using specific primers (Supplementary Table S1 at JXB online) for high-resolution melting (HRM) analysis (Wittwer et al., 2003) using a LightScannerTM (Idaho Technology Inc., USA). The PCR primers for HRM analysis were designed based on the sequence difference at the point-mutation site between Ptd1 and the WT PTD1.
Photoperiod and temperature sensitivity tests
The Ptd1 mutant and ZH11 (WT) plants were treated under different combinations of photoperiod–temperature that encompassed short days (SD, 11/13 h light/dark), long days (LD, 14/10 h light/dark), high temperature (HT, 32/28 °C light/dark with a mean of ~29.8–30.3 °C), and low temperature (LT, 25/23 °C light/ dark with a mean of ~23.9–24.2 °C). The four combinations were SD/HT, LD/HT, SD/LT, and LD/LT. For phenotyping of seedlings, germinated seeds were planted and grown for 8 d in growth chambers set for the different combinations, and for phenotyping over the whole growth period, plants were grown in phytotrons set for the different combinations.
To examine the effects of the photoperiod–temperature combinations on mRNA and protein levels of Ptd1 (in the mutant) and PTD1 (in the WT segregants) in stems at early elongating stage, seedlings were first grown for 45 d in the field under natural conditions in March–April in Guangzhou, China, and then plants were grown for 10 d in phytotrons set at the different combinations. The LD treatment was modified to 14.5/9.5 h light/dark in order to induce a larger difference in expression in response to photoperiod. Three replicate batches of seedlings were used for each treatment.
Vector construction and generation of transgenic plants
Three constructs were designed for mimicking of the mutant. A 5044-bp genomic fragment of PTD1 [WT, including 2456-bp 5´-upstream sequence, 363-bp coding sequence (CDS), 86-bp intron, and 2139-bp 3´-downstream sequences] and a 5043-bp genomic fragment of Ptd1 (mutant, including 2456-bp 5´-upstream sequences, 357-bp CDS, 86-bp intron, and 2144-bp 3´-downstream sequence) were amplified by PCR using the primers shown in Supplementary Table S1. The resultant fragments were subcloned into the pCAMBIA-1300 binary vector via the EcoRI and BamHI cloning sites, resulting in two binary constructs with PTD1 and Ptd1. The third construct expressed a recombinant protein Ptd1::FLAG and was generated by adding a FLAG-encoding tag to Ptd1 using the Omega-PCR method (Chen et al., 2013). To generate site-directed mutations of the conserved Cys codons of 8-CM in PTD1, four modified constructs were generated from the PTD1 vector using Omega-PCR to produce Cys-to-Gly substitutions in the expressed proteins, namely PTD1C102G/C116G, PTD1C56G/C76G, PTD1C31G/C41G, and PTD1C31G/C41G/C102G/C116G. In addition, to examine the role of the C-terminus of PTD1, two constructs expressing PTD1Δ102–120 and PTD1Δ109–120 were prepared in order to produce truncated proteins with deletions of 19 aa and 12 aa of the C-terminus of PTD1, respectively. A construct expressing PTD1::18-aa was also prepared to test the effect of the new 18-aa C-terminus of Ptd1 by fusing it to the full-length PTD1 protein. All the above constructs were transformed into ZH11 using the Agrobacterium-mediated method.
To knockout the PTD1 and Ptd1 allelic genes, a CRISPR/Cas9-knockout construct was generated to target a site in the first exon shared by both PTD1 and Ptd1 (AAGGTGCGCGACGCTGGCGGTGG; the bases in italics are a protospacer adjacent motif, PAM) according to previous protocols (Ma et al., 2015; Ma and Liu, 2016). The resultant construct was transferred into the mutant homozygote (Ptd1/Ptd1) and the WT (PTD1/PTD1) plants by the Agrobacterium-mediated method. The sequencing chromatograms of the targeted mutations were analysed using the DSDecode program (Liu et al., 2015b; Xie et al., 2017).
Genotyping, phenotyping, and growth conditions of the transgenic plants
All transgenic plants were identified by PCR with transgene-specific primer pairs (Supplementary Table S1) followed by sequencing. For phenotyping, transgenic plants and their corresponding WTs were grown in phytotrons, a greenhouse, or in screen-protected fields at the South China Agricultural University in Guangzhou.
RNA extraction and quantitative reverse-transcription PCR
After 10 d of being subjected to the photoperiod-temperature treatments, elongating stems of WT (PTD1/PTD1) and mutant homozygote (Ptd1/Ptd1) plants were collected and ground in liquid nitrogen. The samples were divided into two parts for mRNA and protein detection. Each sample consisted of six plants pooled together, and three replicate samples were analysed. Total RNA extraction was carried out using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. The total RNAs were used to synthesis first-strand cDNA with ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO) following the manufacturer’s instructions. qRT-PCR was performed in a 20-µl reaction using 2× RealStar Green Power Mixture (Genestar, Beijing, China) and a CFX Connect Real-Time PCR Detection System (Bio-Rad) with the qRT-PCR-F/R primers (Supplementary Table S1). Relative expression levels were determined using the 2–△△CT method (Livak and Schmittgen, 2001) with Actin1 (Os03g0718100) as the internal reference.
Western blotting
Total proteins extracted from ground stems of WT and Ptd1 plants were separated by 10% SDS-PAGE (for β-actin protein, MW 42 kDa) or 16% Tris-Tricine SDS-PAGE. Following electrophoretic separation, the protein samples were transferred to polyvinylidene difluoride (PVDF) membranes. After overnight blocking, the membranes were incubated with a solution of primary antibodies diluted in phosphate-buffered saline (1/10 000). Finally, the membranes were stained with a secondary antibody and the bands were detected using a chemiluminescence imaging system (Tanon 5200, Shanghai, China) using enhanced chemiluminescence (ECL, Amersham, USA). The 1–100th aa sequence shared by Ptd1 and PTD1 was used as the antigen to produce an anti-PTD1/Ptd1 polyclonal antibody in rabbits (Beijing Genomics Institute, Shenzhen, China). The mouse anti-β-actin monoclonal antibody and horseradish peroxidase-conjugated goat anti-rabbit IgG (H+L) or anti-mouse IgG (H+L) secondary antibodies (Transgen Biotech Co., Beijing, China) were used in the assays. Ptd1 and PTD1 are expected to have an N-terminus 27-aa extracellular signal peptide, so the molecular weights of the mature proteins (of which the signal peptide is excised) should be 9. 1 kDa and 9. 46 kDa, respectively.
Modeling of the PTD1 protein
The mature protein sequence of PTD1 (excluding the 27-aa signal peptide) was used to construct the 3D structure model using the automatic homology-modeling server SWISS-MODEL (https://www.swissmodel.expasy.org/). The 3D structure of Lc-LTP2 (Protein Data Bank code 2MAL) from lentil (Lens culinaris; Gizatullina et al., 2013) was selected as the template. The model of PTD1 was analysed using the Accelrys Discovery Studio 1.6 software (Accelrys Software Inc., USA).
Results
The Psd1 mutant is photoperiod-thermo-sensitive
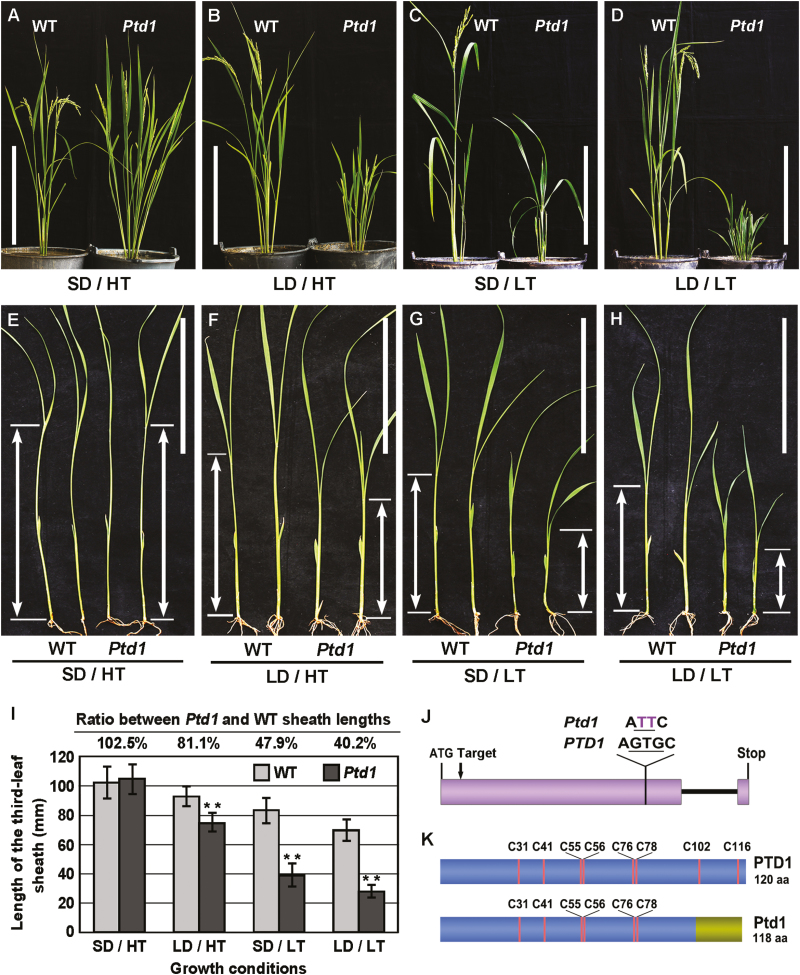
Our previous study had shown that the Psd1 mutant is sensitive to photoperiod: when grown under LD conditions, it displays a severe dwarf phenotype, whilst under SD conditions it is recovered to near-normal plant height (Li et al., 2014). We subsequently observed that the height recovery under SD conditions during a colder season (October–December in Guangzhou, China) was not obvious, suggesting that Psd1 may also be sensitive to temperature. To test this hypothesis, we grew Psd1 plants in two phytotrons set with four combinations of high or low temperatures (HT, LT) and long or short days (LD, SD). The results showed that under SD/HT conditions, the plant height of Psd1 was similar to that of the WT (Fig. 1A), while under LD/HT, SD/LT, and LD/LT conditions it had obvious dwarf phenotypes, with the mostly severe dwarfism under LD/LT (Fig. 1B–D). In contrast, the height of the WT was increased slightly under LT and/or LD conditions (Fig. 1A–D).
Fig. 1.
The rice Photoperiod-thermo-sensitive dwarfism 1 (Ptd1) mutant and the causal gene. (A–D) Plant height of Ptd1 and wild-type (WT) plants after 90 d of photoperiod–temperature treatments: SD, short day; LD, long day; HT, high temperature; LT, low temperature. Scale bars are 30 cm. (E–I) Length of the third leaf sheath of Ptd1 and WT seedlings after 8 d of treatment. The arrows indicate the lengths of the sheaths. Scale bars are 10 cm. (I) Length of the third leaf sheaths presented as means (±SD), n=20. Significant differences compared with the WT were determined using Student’s t-test: **P<0.01. (J) The gene structures of PTD1 (WT allele) and Ptd1 (mutant allele). PTD1 contains two exons (shaded region, the UTRs are not shown) and one intron (black line). A point mutation (GTG to TT) in the first exon of PTD1 results in Ptd1. ‘Target’ indicates the site targeted by CRISPR/Cas9. (K) The protein structures of PTD1 and Ptd1. The positions of the eight conserved Cys residues are indicated. The region at the right-hand end of Ptd1 is the newly-formed 18-amino acid C-terminus that results from the frame-shifted mutation. (This figure is available in color at JXB online.)
To provide a quantitative assessment of the effects, the length of the sheath of the third leaves of seedlings was measured after 8 d of treatment under the various conditions. The growth of the Psd1 seedlings was the same as the WT under SD/HT (Fig. 1E, I), but they showed significant dwarf phenotypes under LD/HT, SD/LT, and LD/LT conditions, with the sheath lengths reduced to 81.1%, 47.9%, and 40.2% of the WT, respectively (Fig. 1F–I). These results indicated that both LD and LT conditions contributed to the growth inhibition of Psd1, and that the temperature effect was greater than the day-length effect (Fig. 1I). Since Psd1 was sensitive to both photoperiod and temperature, we hereafter rename it as Photoperiod-thermo-sensitive dwarfism1 (Ptd1).
A point-mutation in Os01g0822900 causes the dominant Ptd phenotype
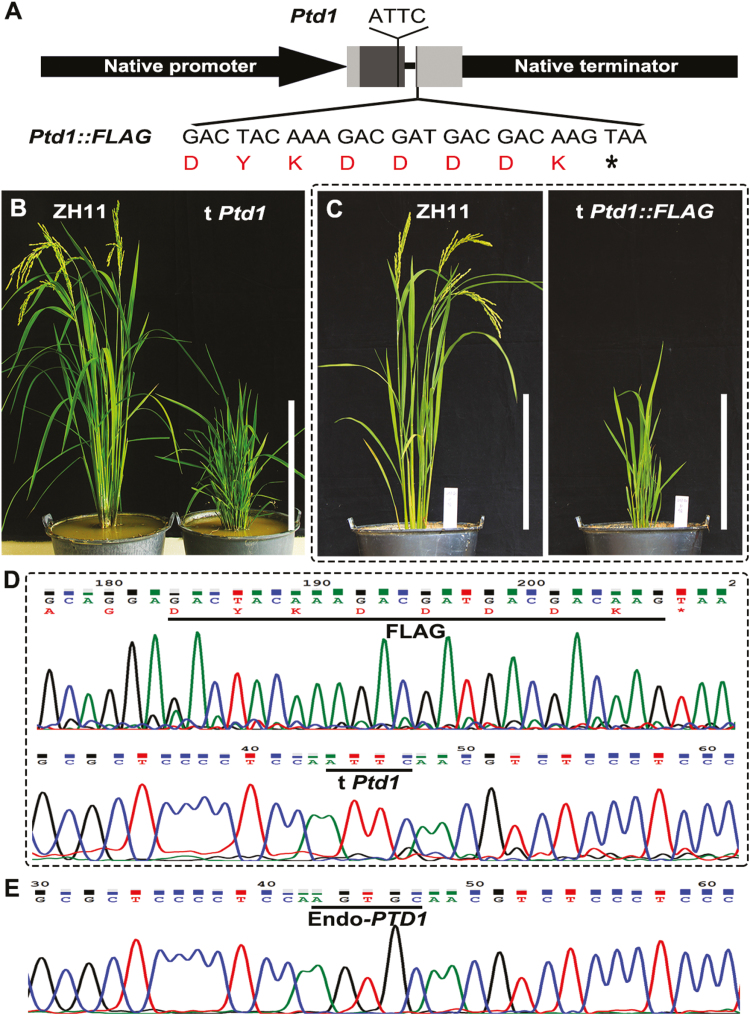
We have previously mapped Ptd1 to a candidate gene Os01g0822900 (Supplementary Fig. S1), which encodes a putative nsLTP. The allele of Os01g0822900 in the Ptd1 mutant carries a single-nucleotide substitution and a single-nucleotide deletion, thus leading to a frame-shift from the 101st codon (Li et al., 2014) (Fig. 1J, K). To verify the candidate gene in our current study, we prepared a binary construct of Ptd1 with the genomic DNA of the Ptd1 allele (with full length of 5043 bp; see methods) and another construct Ptd1::FLAG fused with the FLAG tag (Fig. 2A), and transferred them into the WT ZH11. If the Ptd1 transgene is dominantly functional, Ptd1-transgenic plants grown in LD and/or LT conditions would show the dwarf phenotype. As expected, 9 of the 12 independent transgenic lines obtained (grown in LD conditions) mimicked the dwarf phenotype of Ptd1 (Fig. 2B). Similarly, all eight transgenic lines with Ptd1::FLAG also displayed the dwarf phenotype of Ptd1 (Fig. 2C–E). The mutant allele of Os01g0822900 was thus confirmed to be the causal gene of Ptd1.
Fig. 2.
Mimicking of the Ptd1 mutant in rice. (A) Constructs used for mutant-mimicking transformation. Ptd1 or Ptd1::FLAG was controlled under the native promoter and terminator of PTD1. The two shaded boxes represent the exons, with dark grey parts being the CDS and the light grey parts being the 5´ and 3´ UTRs. The mutation site (ATTC) of Ptd1 is marked, and the codons and amino acids of the fused FLAG are shown. (B, C) Phenotypes of the recipient wild-type ZH11 and the transgenic plants (t Ptd1 and t Ptd1::FLAG) grown under natural long-day conditions (~13.5–14.4 h and ~25–28 °C). Scale bars are 30 cm. (D, E) Sequencing verification of the Ptd1::FLAG-transgenic plants. The transgene Ptd1 (characterized by ATTC) and the fused FLAG sequences are shown in (D), and the endogenous PTD1 (Endo-PTD1, characterized by AGTGC) is shown in (E). (This figure is available in color at JXB online.)
Since PTD1 is predicted to be a nsLTP, we further tested the lipid-binding ability of PTD1/Ptd1 with a fluorescence-labeled lipid 1-pyrenedodecanoic fatty acid (p-96), using the method described by Zhang et al (2010). As expected, the recombinant PTD1 protein showed a high level of lipid-binding activity in vitro, more than twice that of the positive control, OsC6, while the recombinant Ptd1 displayed a much weaker activity, about half of that of OsC6 (Supplementary Fig. S2).
Ptd1 expression and its protein accumulation are affected by photoperiod and temperature, and account for the Ptd phenotype
It was possible that the variations in plant height of Ptd1 may have resulted from the expression of Ptd1 per se in response to the variations in photoperiod and temperature, or that the mutant gene Ptd1 may have hampered the functions of other related gene(s) sensitive to photoperiod and temperature. Since the expression of nsLTP family genes displays complex patterns in response to various environmental factors (Sohal et al., 1999; Kiełbowicz-Matuk et al., 2008; Liu et al., 2015a), we speculated that mRNA expression or protein accumulation of Ptd1 may respond to alterations in temperature and photoperiod. Hence, we detected mRNA and protein levels of Ptd1 and PTD1 (in WT segregants) in early elongating stems sampled from plants after treatment under the different photoperiod–temperature combinations.
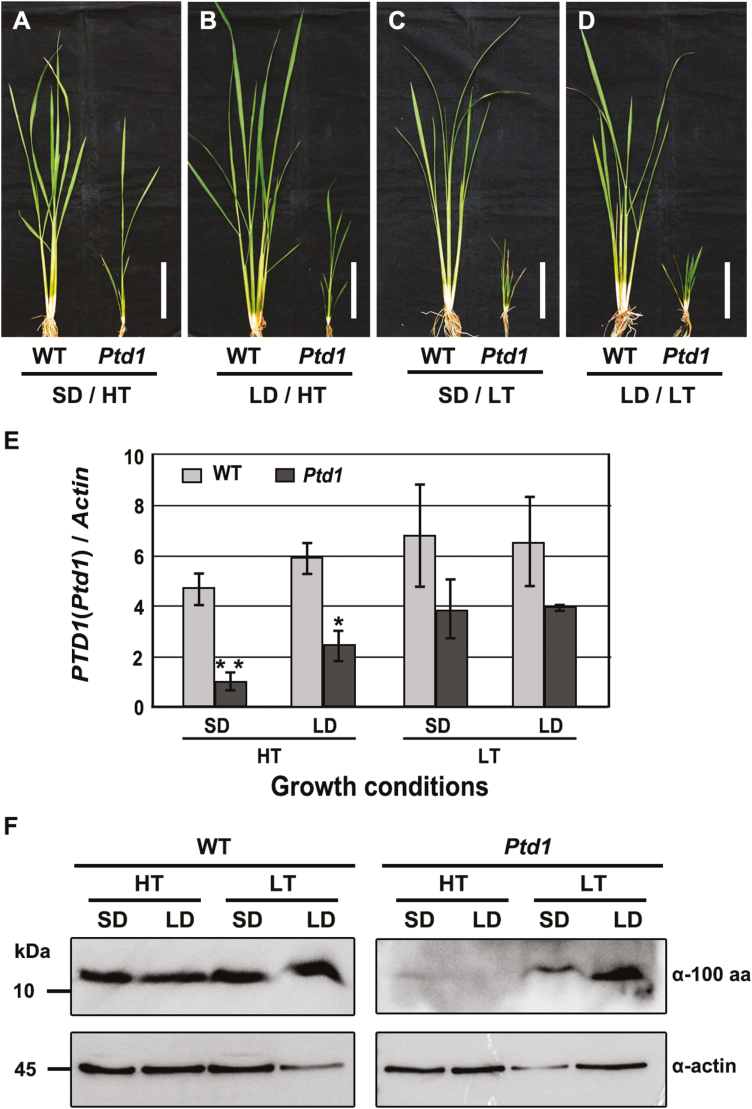
Ptd1 and the WT were first grown under natural LD/LT conditions for 45 d (~13.5–14 h daylength, mean temperature ~23–25 °C), which resulted in Ptd1 plants that were <20 cm in height. These plants (which were in the early stem-elongation stage) were then treated for 10 d in phytotrons under the different photoperiod–temperature conditions (with LD=14.5/9 h light/dark in this case). The Ptd1 plants showed distinct growth responses to the four different sets of conditions. Under SD/HT, the growth of Ptd1 was considerable relieved (Fig. 3A) and under LD/HT it was slightly relieved (Fig. 3B); however, severe growth inhibition remained under both SD/LT and LD/LT conditions (Fig. 3C, D). In contrast, the WT plants showed no significant changes in growth under any of the four sets of conditions (Fig. 3A–D). These results were consistent with those obtained under the continuous photoperiod–temperature treatments (Fig. 1A–I).
Fig. 3.
Alterations of mRNA expression and protein accumulation in the rice Ptd1 and wild-type plants in response to different photoperiod–temperature treatments. (A–D) Height of the Ptd1 and wild-type (WT) plants grown under natural long-day/low-temperature conditions for 45 d followed by 10 d of treatment under different photoperiod-temperature combinations: SD, short day; LD, long day; HT, high temperature; LT, low temperature. Scale bars are 10 cm. Stems of the treated plants were sampled for analysis of mRNA expression and protein accumulation. (E) Expression of PTD1 and Ptd1 in the WT and Ptd1 mutant. Data are means (±SE) of three biological replicates. (F) Accumulation of PTD1 and Ptd1 proteins in the WT and Ptd1 mutant. α-100 aa, anti-PTD1/Ptd1 antibody; α-actin, anti-actin antibody. (This figure is available in color at JXB online.)
At the end of the 10-d treatments, we sampled the stems for mRNA and protein analyses. Under all the photoperiod–temperature combinations, the changes in expression of Ptd1 were similar to those of the WT gene PTD1, but the mRNA levels of Ptd1 were lower than those of PTD1 (Fig. 3E). However, comparison among the photoperiod–temperature treatments in Ptd1 indicated that the mRNA levels under LT were higher than those under HT, and that the level under SD/HT was the lowest. These results indicated that the degree of growth inhibition increased in proportion to the mRNA levels of Ptd1 (Fig. 3E).
We next examined the accumulation of Ptd1 and PTD1 proteins in the stems. An antibody was produced to detect both proteins using the 1–100th aa common sequence shared by Ptd1 and PTD1 as the antigen. The specificity of the antibody was verified by western blotting, using protein samples expressed from the prokaryotic system. Specific bands were detected in the samples containing PTD1 and Ptd1, but not in with the control nsLTP protein OsC6 (Zhang et al, 2010), or in the negative control sample (Supplementary Fig. S3). These results indicated that the antibody was specific to PTD1 and Ptd1, and hence we used it to detect the protein levels in our tissue samples. The results showed that accumulation of Ptd1 under LT conditions was higher than that under HT conditions, with the highest accumulation being observed under LD/LT and a dramatic decrease being observed under HT (Fig. 3F). This accumulation pattern was in agreement with the mRNA expression pattern, and higher levels were associated with more severe dwarfism in the Ptd1 plants. In contrast, the accumulation of the WT PTD1 protein remained relatively constant under the different conditions (Fig. 3F). These results may explain the complete or partial restoration of the plant height of Ptd1 under HT (Fig. 1A–I). Further, under the LT conditions, SD depressed accumulation of Ptd1 to a degree and slightly relieved the inhibition of growth. When the mutant had low Ptd1 accumulation under HT conditions, the effect of photoperiod on Ptd1 accumulation was not obvious (Fig. 3F), and the mutant plants were recovered to almost normal under SD treatments. Hence, temperature is the major factor controlling the growth of Ptd1 plants, and photoperiod acts as a minor factor that further influences their growth.
Knockout mutants of PTD1 and Ptd1 display a normal growth phenotype
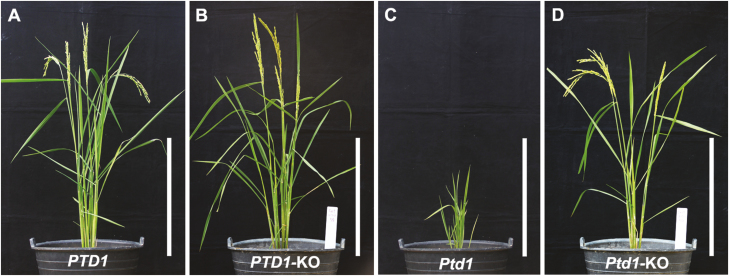
To confirm the function of Ptd1 and to determine whether PTD1 is involved in the control of plant height, we performed CRISPR/Cas9 gene editing (Ma et al., 2015) by targeting the PTD1 and Ptd1 alleles. A CRISPR/Cas9 target site was designed near the start codon in the first exon of PTD1 and Ptd1 (Fig. 1J). We obtained five and three independent knockout lines from the targeted Ptd1 and PTD1 lines, respectively, with biallelic or homozygous mutations (all causing frame-shifts) (Supplementary Table S2). All the knockout plants showed normal growth from the T0 to T2 generations, similar to the WT (Fig. 4A–D). Taken together, these results indicate that the primary function of PTD1 may not involve in plant growth, or it may be masked by gene redundancy, and it is evident that Ptd1 is a dominant gain-of-function mutant.
Fig. 4.
Functional knockout of rice PTD1 and Ptd1 by CRISPR/Cas9. Phenotypes are shown of the wild-type (PTD1), the Ptd1 mutant, and their knockout lines (-KO). Scale bars are 30 cm. The target genotypes and sequencing chromatograms of the knockout plants are shown in Supplementary Table S2. (This figure is available in color at JXB online.)
The Ptd1 phenotype is independent to the protein C-terminus of PTD1
The frame-shift mutation of the Ptd1 gene results in a truncated protein of 118 aa with a new 18-aa C-terminus (Fig. 1K). In order to determine whether the additional C-terminal alteration contributes to the gain-of-function, we designed constructs to produce modified PTD1, namely two with truncations of the last 19 aa (PTD1Δ102–120) and 12 aa (PTD1Δ109–120) off the C-terminus, and one recombinant PTD1 fusion with an additional 18-aa frame-shifted C-terminus (PTD1::18 aa) (Fig. 5A–D). These constructs were transferred into the WT (ZH11), and the resultant transgenic lines were planted through to the T1 generation (three lines for PTD1Δ102–120, five for PTD1Δ109–120, and five for PTD1::18 aa) and to the T2 generation (two lines for PTD1Δ102–120, two for PTD1Δ109–120, and four for PTD1::18 aa) for phenotyping. We found that all the transgenic lines had similar plant height to the WT (Fig. 5B–D). We therefore deduced that the dominant gain-of-function of Ptd1 was not due to either the new frame-shifted C-terminus or merely the loss of the WT C-terminus. Instead, we concluded that it may have been caused by a specific conformation present in the Ptd1 protein.
Fig. 5.
Transgenic rice plants with modified PTD1 constructs encoding mutated C termini. (A) The recipient wild-type ZH11 carries a PTD1 gene encoding an endogenous wild-type protein (Endo-PTD1). (B–D) Schematic protein structures from the modified PTD1-constructs encoding different C termini, and their corresponding transgenic plants. The vertical lines indicate the positions of the conserved Cys residues in PTD1 and the mutant products. The proteins PTD1Δ102–120 and PTD1Δ109–120 have deletions of the segments of 102–120th aa and 109–120th aa of PTD1, respectively, and PTD1::18 aa is a recombinant protein fused with the mutant 18 aa-C terminus of Ptd1 (shaded box at the right-hand end). Scale bars are 30 cm. (This figure is available in color at JXB online.)
Loss of the specific disulfide bonds in the 8-CM of PTD1 causes the Ptd1 phenotype
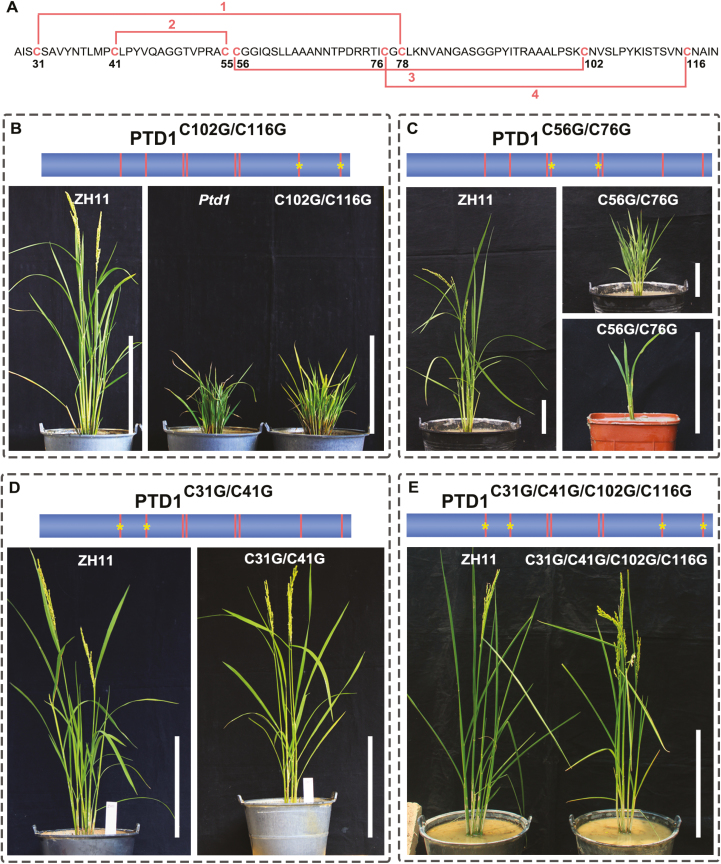
NsLTPs are characterized by the presence of a conserved 8-CM motif capable of forming four disulfide bonds, which play an important role in maintaining the particular tertiary structure of these proteins (Kader, 1996; José-Estanyol et al., 2004; Salminen et al., 2016). The PTD1 protein shares the 8-CM motif, which is predicted to form four disulfide bridges in the pattern Cys31-Cys78, Cys41-Cys55, Cys56-Cys102, and Cys76-Cys116 (Boutrot et al., 2008) (Fig. 6A). However, Ptd1 loses the Cys102 and Cys116 residues and the disulfide bonds Cys56-Cys102 and Cys76-Cys116 (Fig. 1K). We speculated that the acquired function of Ptd1 may result from the destruction of these disulfide bonds. We therefore designed four constructs carrying PTD1 (driven by its native promoter) with modified codons to change the conserved Cys to Gly in the 8-CM motif, and transferred them into the WT (ZH11). The constructs encoding PTD1C102G/C116G and PTD1C56G/C76G were designed to destroy the last two disulfide bonds (Fig. 6A–C) to mimic the effect of Ptd1, and the constructs encoding PTD1C31G/C41G and PTD1C31G/C41G/C102G/C116G were designed to destroy the first two and all four disulfide bonds, respectively, (Fig. 6A, D, E) with the aim of detecting the biological effects of these bonds.
Fig. 6.
Transgenic rice plants carrying modified PTD1 constructs with codon substitutions for the conserved cysteine sites. (A) Schematic diagram of the four disulfide bonds (lines 1–4) formed by the eight-cysteine motif in PTD1. The positions of the bonds are numbered. The 27-aa signal peptide in the N terminus is not shown. (B, C) T0 transgenic plants carrying modified PTD1 without both the 3rd and 4th disulfide bonds due to the Cys-to-Gly substitutions of PTD1C102G/C116G (B) and PTD1C56G/C76G (C). The wild-type ZH11 is also shown. Two representative transgenic plants are shown in (C). The plants were grown under natural long-day/high-temperature (LD/HT) conditions (B) and in short-day /low-temperature (SD/LT) conditions in a greenhouse (C). (D) T1 transgenic plants carrying modified PTD1 without both the 1st and 2nd disulfide bonds due to the Cys-to-Gly substitutions of PTD1C31G/C41G. (E) T0 transgenic plants carrying modified PTD1 without all four of the disulfide bonds due to the Cys-to-Gly substitutions of PTD1C31G/C41G/C102G/C116G. The plants in (D, E) were grown under natural LD/LT conditions. In the schematic diagrams above the images, the vertical lines indicate the positions of the conserved Cys residues in PTD1, and * indicates the positions of the lost Cys residues. The recipient is ZH11. Scale bars are 30 cm in (B, D, E), and 10 cm in (C). The sequencing verification of the transgenic plants is shown in Supplementary Fig. S4. (This figure is available in color at JXB online.)
A total of 48 independent T0 transgenic plants expressing PTD1C102G/C116G were obtained, and all of them presented a Ptd1-like dwarf phenotype (Fig. 6B). Similarly, all the 20 independent T0 transgenic plants expressing PTD1C56G/C76G obtained displayed severe reduction of growth (Fig. 6C). These results demonstrated that the loss of the third and fourth disulfide bonds caused the dominant gain-of-function of Ptd1 and the inhibition of growth.
We further investigated the effects of the first two and all four disulfide bonds in PTD1. However, all the transgenic plants expressing PTD1C31G/C41G (lacking the first two bonds) or PTD1C31G/C41G/C102G/C116G (lacking all four bonds) showed normal growth, similar to the WT plants (Fig. 6D, E). Taken together, it is reasonable to speculate that the acquired function of Ptd1 depends on a specific protein conformation that results from the loss of the last two disulfide bonds and the retention of the first two of the 8-CM motif in PTD1.
The Ptd1 phenotype requires a specific protein conformation
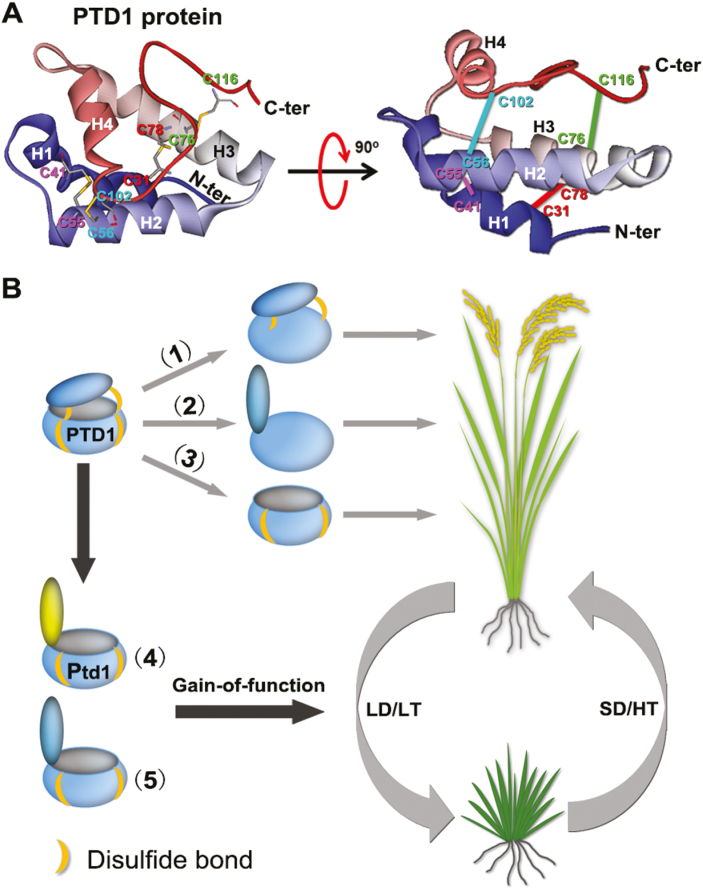
In order to explore the tertiary structure of PTD1/Ptd1, we performed protein structure homology modeling. The amino acid sequence of PTD1 excluding the 27-aa signal peptide at the N-terminus was used to search for template homologous proteins in the SWISS-MODEL database. We found a homolog, Lc-LTP2, from Lens culinaris (PDB ID: 2MAL) sharing 52.17% identity with PTD1, and therefore its 3D structure was selected as a template to predict that of PTD1.As shown in Fig. 7A, PTD1 forms a central hydrophobic cavity that is surrounded by four α-helices and covered by a C-terminal lid. The structure of PTD1 is consolidated by four disulfide linkages, of which Cys31-Cys78 and Cys41-Cys55 are for fixing the pocket, while Cys56-Cys102 and Cys76-Cys116 are for screwing between the lid and the pocket. In Ptd1, however, the last two disulfide bonds (Cys56-Cys102 and Cys76-Cys116) are destroyed, which may lead to impaired fixation between the C-terminal lid and the hydrophobic cavity (Fig. 7B). Given that all the types of transgenic plants that harbored the modified PTD1 without the pocket structure (PTD1C31G/C41G and PTD1C31G/C41G/C102G/C116G) (Figs 6D, E, 7B) or the C-terminal lid (PTD1Δ102–120) (Figs 5B, 7B) presented a normal phenotype, we speculate that the gain-of-function of Ptd1 requires a specific protein conformation with a hydrophobic pocket and an open C-terminal lid with a suitable length.
Fig. 7.
Gain-of-function of rice Ptd1 may be due to the specific conformation of the protein. (A) Predicted 3D structure of PTD1 (excluding the 27-aa signal peptide in the N terminus). Left, top view. The positions of the conserved Cys residues are indicated by ‘C’ followed by a number. The pairs C31–C78, C41–C55, C56–C102, and C76–C116 each form a specific disulfide linkage (shown by the connecting bars). Right, side view. The straight lines correspond to the disulfide linkages shown in the top-view. H1–H4, indicate the four alpha helixes. C-ter and N-ter are C and N termini, respectively. (B) Gain-of-function of Ptd1 may be due to the specific protein conformation. The PTD1 protein forms a hydrophobic pocket structure with a C-terminal lid, consolidated by four disulfide bonds. Loss of the lower two (1) or all four (2) disulfide bonds, or loss of the C terminus (3) has no biological effect on plant growth (i.e. no inhibition). In contrast, the mutated protein Ptd1 (4), which is expressed under low-temperature (LT) and/or long-day (LD) conditions, loses the upper two disulfide bonds that screw the lid and the pocket, but retains the lower two disulfide bonds for pocket-fixing. This protein conformation may form a molecular ‘catcher’ or ‘trap’ and thus the protein gains a biological function for growth inhibition. The conformation in (5) represents the artificially mutated PTD1 forms in the transgenic plants (PTD1C102G/C116G and PTD1C56G/C76G; Fig. 6B, C). Under high-temperature (HT) and short-day (SD) conditions, the expression of Ptd1 is at relatively low levels (Fig. 3E, F) and hence plant growth is recovered. (This figure is available in color at JXB online.)
Discussion
NsLTPs are a class of small proteins that are widely distributed in higher plants, and their abundance can be as high as 4% of the total soluble proteins (Liu et al., 2015a). Since the first nsLTP was characterized, considerable efforts have been made to determine their biological functions. In most instances, it can be hard to characterize any abnormal phenotype in nsLTP knockdown or knockout mutants due to functional redundancy among the nsLTP family members (Salminen et al., 2016). However, when a phenotype is attributed to a mutated nsLTP gene, care needs to be taken to determine whether the mutant trait is caused by loss-of-function of the WT gene or by gain-of-function of the mutant allele. Only the effect(s) due to loss-of-function or overexpression of the WT gene will reflect the original role of the nsLTP gene.
The expression of Ptd1 (and PTD1) was affected by day length and temperature (Fig. 3E), and thus the dominant Ptd1 trait was photoperiod-thermo-sensitive. We initially assumed that the original function of PTD1 might somehow be related to the regulation of plant height, probably by a mechanism different to that of the Ptd1 allele. However, PTD1-knockout plants showed a normal phenotype (Fig. 4). Despite this, it is not possible to conclude that PTD1 is not involved in growth regulation, given that there are many (52) nsLTP members in rice (Boutrot et al., 2008), and gene redundancy may exist.
Ptd1 and the PTD1 only differed in the 3´-end of the coding region and their mRNA expression trends were similar in response to the photoperiod–temperature treatments, but the mRNA levels of Ptd1 were lower than those of PTD1 under all four treatment combinations (Fig. 3E). This may have been due to the common nonsense-mediated mRNA decay mechanism (Baker and Parker, 2004) since the Ptd1 mRNA carried a point-mutation. As regards the mechanism by which the Ptd1 plants showed different phenotypes under LD/LT and SD/HT conditions, we propose that Ptd1 functions at the protein level to inhibit growth, with the conformation of the Ptd1 protein potentially being unstable under the HT treatment and prone to degradation (Fig. 3F). Synergistically with the changes in mRNA expression in response to photoperiod and temperature and with the nonsense-mediated mRNA decay mechanism acting on Ptd1 mRNA, the level of functional Ptd1 protein was highest under LD/LT conditions and lowest under SD/HT. Thus, the Ptd1 plants showed severe growth inhibition under LD/LT, and this inhibition was relieved under SD/HT.
The members of nsLTP family share a conserved 8-CM motif capable of forming four disulfide bonds, which are important for shaping the functional structures of the proteins. The 8-CM shows no catalytic activity in plants (Mullaney and Ullah, 2005). However, the disulfide bonds confer stability to the hydrophobic cavity, a structure for binding various hydrophobic compounds (Douliez et al., 2000). Our results indicated that the loss of the 3rd and 4th disulfide bonds in the Ptd1 protein caused the gain-of-function for growth inhibition (Fig. 6B, C). However, this alone was not sufficient: the existence of the 1st and 2nd disulfide bonds (Fig. 6D, E) and a suitable C-terminal tail (Fig. 5B) were also indispensable, as summarized in Fig. 7B. This implies that the gain-of-function of Ptd1 depends on a relatively open ‘pocket’ rather than no ‘pocket’ at all (Fig. 7B). Hence, we suggest that Ptd1 may acquire its new function as a result of an alteration of its conformation that makes it capable of interacting with specific ligands or allows it to act as a molecular ‘catcher’ or ‘trap’ for key factor(s) of particular pathways required for cell division and elongation, with the end result being growth inhibition. In this sense, Ptd1 is a neomorph. It was notable that Ptd1 seedlings showed distinct growth responses after only 8 d of photoperiod–temperature treatment (Fig. 1E–I), suggesting that Ptd1 may be a strong ‘catcher’. Further studies of the ligand/protein-binding of Ptd1 are required to provide a better understanding of the precise mechanisms by which it regulates cell proliferation and elongation.
The conformation of a protein is fundamental to its function. In molecular engineering, site-directed mutagenesis combined with knowledge-based prediction of protein conformation is a powerful tool for designing such things as novel enzymes and antibodies (Blundell et al., 1987). In this study, we successfully engineered a nsLTP protein to create a new function by making slight modifications in the 8-CM motif (Fig. 6B, C). As a conserved structural motif, 8-CM has been detected in a large number of proteins with various functions in the plant kingdom (José-Estanyol et al., 2004). Our findings potentially provide new directions for understanding and engineering this group of proteins.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The CDS sequence of PTD1 (Os01g0822900).
Fig. S2. Recombinant PTD1 protein displays lipid-binding activity.
Fig. S3. Verification of the specificity of antibody α-PTD11-100aa by western blotting.
Fig. S4. Sequencing verification of the transgenic plants carrying modified PTD1 constructs with codon substitutions for the conserved cysteine sites.
Table S1. List of primers used in this study.
Table S2. Genotypes and sequencing chromatograms of the CRISPR/Cas9 knockout mutants of PTD1 and Ptd1.
Acknowledgements
We thank Prof. Qingyin Zeng at the Institute of Botany, Chinese Academy of Sciences, Beijing, China, for his kind assistance with protein modeling and protein-structure analysis using the Accelrys Discovery Studio 1.6 software. This work was supported by grants from the Ministry of Agriculture of China (2016ZX08010001) and the National Natural Science Foundation of China (31271301) for YC.
References
- Ambrose C, DeBono A, Wasteneys G. 2013. Cell geometry guides the dynamic targeting of apoplastic GPI-linked lipid transfer protein to cell wall elements and cell borders in Arabidopsis thaliana. PLoS ONE 8, e81215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KE, Parker R. 2004. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Current Opinion in Cell Biology 16, 293–299. [DOI] [PubMed] [Google Scholar]
- Blundell TL, Sibanda BL, Sternberg MJ, Thornton JM. 1987. Knowledge-based prediction of protein structures and the design of novel molecules. Nature 326, 347–352. [DOI] [PubMed] [Google Scholar]
- Boutrot F, Chantret N, Gautier MF. 2008. Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics 9, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Kieslich CA, Morikis D, Kim SC, Lord EM. 2009. A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. The Plant Cell 21, 3902–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang F, Wang X, Liu YG. 2013. Robust one-tube Ω-PCR strategy accelerates precise sequence modification of plasmids for functional genomics. Plant & Cell Physiology 54, 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. 1996. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiology 111, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBono A, Yeats TH, Rose JK, Bird D, Jetter R, Kunst L, Samuels L. 2009. Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. The Plant Cell 21, 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douliez J, Michon T, Elmorjani K, Marion D. 2000. Mini review: structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. Journal of Cereal Science 32, 1–20. [Google Scholar]
- Evenson RE, Gollin D. 2003. Assessing the impact of the green revolution, 1960 to 2000. Science 300, 758–762. [DOI] [PubMed] [Google Scholar]
- Gizatullina AK, Finkina EI, Mineev KS, Melnikova DN, Bogdanov IV, Telezhinskaya IN, Balandin SV, Shenkarev ZO, Arseniev AS, Ovchinnikova TV. 2013. Recombinant production and solution structure of lipid transfer protein from lentil Lens culinaris. Biochemical and Biophysical Research Communications 439, 427–432. [DOI] [PubMed] [Google Scholar]
- José-Estanyol M, Gomis-Rüth FX, Puigdomènech P. 2004. The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiology and Biochemistry 42, 355–365. [DOI] [PubMed] [Google Scholar]
- Jung HW, Kim KD, Hwang BK. 2005. Identification of pathogen-responsive regions in the promoter of a pepper lipid transfer protein gene (CALTPI) and the enhanced resistance of the CALTPI transgenic Arabidopsis against pathogen and environmental stresses. Planta 221, 361–373. [DOI] [PubMed] [Google Scholar]
- Kader JC. 1996. Lipid-transfer proteins in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 627–654. [DOI] [PubMed] [Google Scholar]
- Kielbowicz-Matuk A, Rey P, Rorat T. 2008. The organ-dependent abundance of a Solanum lipid transfer protein is up-regulated upon osmotic constraints and associated with cold acclimation ability. Journal of Experimental Botany 59, 2191–2203. [DOI] [PubMed] [Google Scholar]
- Lee SB, Go YS, Bae HJ, Park JH, Cho SH, Cho HJ, Lee DS, Park OK, Hwang I, Suh MC. 2009. Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiology 150, 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Xia J, Xu Y, Zhao X, Liu YG, Chen Y. 2014. Characterization and genetic mapping of a Photoperiod-sensitive dwarf 1 locus in rice (Oryza sativa L.). Theoretical and Applied Genetics 127, 241–250. [DOI] [PubMed] [Google Scholar]
- Liu F, Zhang X, Lu C, Zeng X, Li Y, Fu D, Wu G. 2015a Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. Journal of Experimental Botany 66, 5663–5681. [DOI] [PubMed] [Google Scholar]
- Liu W, Xie X, Ma X, Li J, Chen J, Liu YG. 2015b DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Molecular Plant 8, 1431–1433. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma X, Liu YG. 2016. CRISPR/Cas9-based multiplex genome editing in monocot and dicot plants. Current Protocols in Molecular Biology 115, 31.6.1–31.6.21. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, et al. . 2015. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. 2002. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419, 399–403. [DOI] [PubMed] [Google Scholar]
- Mori M, Nomura T, Ooka H, et al. . 2002. Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiology 130, 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney EJ, Ullah AH. 2005. Conservation of cysteine residues in fungal histidine acid phytases. Biochemical and Biophysical Research Communications 328, 404–408. [DOI] [PubMed] [Google Scholar]
- Nichols JW. 1987. Binding of fluorescent-labeled phosphatidylcholine to rat liver nonspecific lipid transfer protein. The Journal of Biological Chemistry 262, 14172–14177. [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BA, Fasolino A, Hilbers CW, Derksen J, Mariani C. 2005. Lipid transfer proteins enhance cell wall extension in tobacco. The Plant Cell 17, 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel D. 1996. Green revolution agriculture and chemical hazards. The Science of the Total Environment 188, S86–S98. [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Datta S, Persak H. 2014. Salt stress in Arabidopsis: lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Molecular Plant 7, 722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen TA, Blomqvist K, Edqvist J. 2016. Lipid transfer proteins: classification, nomenclature, structure, and function. Planta 244, 971–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, et al. . 2002. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. [DOI] [PubMed] [Google Scholar]
- Shinichiro W, Mitsuhiro Y. 1986. Purification and characterization of a non-specific lipid transfer protein from germinated castor bean endosperms which transfers phospholipids and galactolipids. Biochimica et Biophysica Acta - Lipids and Lipid Metabolism 876, 116–123. [Google Scholar]
- Sohal AK, Pallas JA, Jenkins GI. 1999. The promoter of a Brassica napus lipid transfer protein gene is active in a range of tissues and stimulated by light and viral infection in transgenic Arabidopsis. Plant Molecular Biology 41, 75–87. [DOI] [PubMed] [Google Scholar]
- Tsuboi S, Osafune T, Tsugeki R, Nishimura M, Yamada M. 1992. Nonspecific lipid transfer protein in castor bean cotyledon cells: subcellular localization and a possible role in lipid metabolism. Journal of Biochemistry 111, 500–508. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, et al. . 2005. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. 2003. High-resolution genotyping by amplicon melting analysis using LCGreen. Clinical Chemistry 49, 853–860. [DOI] [PubMed] [Google Scholar]
- Xie X, Ma X, Zhu Q, Zeng D, Li G, Liu YG. 2017. CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Molecular Plant 10, 1246–1249. [DOI] [PubMed] [Google Scholar]
- Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D. 2010. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiology 154, 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, et al. . 2013. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.