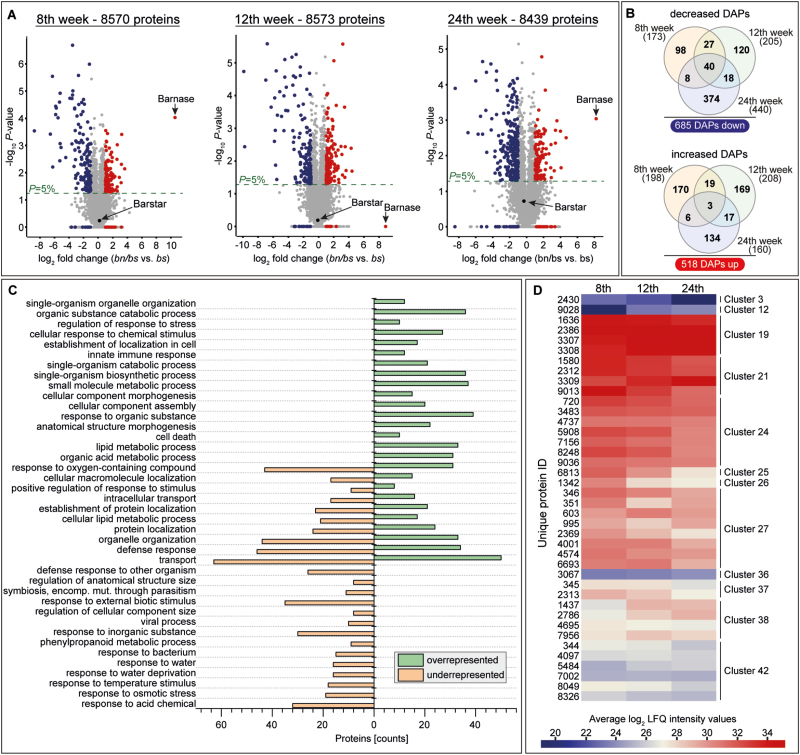
Fig. 5.
Identification of differentially accumulated proteins (DAPs) between roots of transgenic plants of Taraxacum koksaghyz that either exude latex (barstar, bs) or do not exude latex (barnase, bn/bs). Roots were sampled when plants were 8, 12, and 24 weeks old. (A) The protein ratios are displayed as volcano plots: increased DAPs are shown in blue, decreased ones are shown in red. Proteins or protein groups that showed a log2 ratio of >1 or <–1 and *P <0.05 were considered as differentially accumulated. In addition, proteins that were not detectable (invalid log2 ratio) or that were almost absent in one of the genotypes (quantified only for one out of four replicates, thus not allowing for the generation of a P-value) were also taken into account; however, a prerequisite for this was the detection of these proteins in the other comparative genotype in at least three out of four replicates. (B) Venn diagrams illustrating the distribution of the dataset across the three time-points. (C) GO term analysis of the detected DAPs. The biological processes shown met the criteria that at least eight proteins had to be counted and have a calculated P-value <0.05. (D) Heatmap illustrating the temporal protein accumulation patterns in the near-isogenic line (NIL) proteome dataset for the 40 proteins that were detected as differentially less accumulated at all three time-points. The corresponding cluster allocations are shown to the right. See Supplementary Fig. S4 and Supplementary Table S5.