Highlights
-
•
Tripartite symbiosis between plants, their beneficial microbes and toxigenic microbes exists.
-
•
The toxigenic microbes negatively impact crop productivity and constitute public health threat.
-
•
Metabolomics of this complex tripartite interaction in tandem could generate new biomarkers useful in plant biotechnology.
-
•
Future biomarker application in novel crop protection and phytopathogen monitoring/diagnostic approaches were discussed.
Keywords: Beneficial microbe, Crop protection, Metabolomics, Phytopathogen, Plant-microbe interaction
Abstract
Phytopathogens from the Alternaria sp., Fusarium sp., Penicillium sp., and Pseudomonas sp. and their toxigenic metabolites - alternariol, fumonisin, citrinin, and coronatine respectively, negatively impact crop yields and sales by eliciting plant diseases and/or causing human and veterinary toxicoses upon the consumption of contaminated food. These phytopathogens and their associated toxins, however, are present and most likely in undetectable concentrations pre-harvest and post-harvest of many major staple crops. Metabolomic approaches have been used extensively for better characterizing and diagnosing human disease, plant disease and, their etiological agents. Their use in agro-industrial research focusing specifically on tripartite (plant - toxicogenic microbe - beneficial microbe) interactions is, however, limited. Since new approaches for eradicating food-borne pathogens, increasing crop productivity and improving agro-international trade are being sought worldwide, the consequent integration of metabolomic approaches and perspectives in crop protection strategies for better understanding plant - toxicogenic microbe - beneficial microbe interaction in tandem is discussed.
1. Introduction
Food shortage and insecurity are still global issues because of the ever-increasing human population, and the corresponding increased demands this places on farming. Many agro-industrial plant-borne fungal and bacterial pathogens (e.g. Alternaria sp., Burkholderia sp., Fusarium sp., Penicillium sp., Pseudomonas sp., Rhizoctonia sp., and Xanthomonas sp.), produce bioactive metabolites (alternariol, toxoflavin, fumonisin, citrinin, coronatine, RS-toxin, and albicidin respectively), that compromise the quality and usability of harvested crops, subsequently negatively impacting human and animal health. These phytopathogens are mostly associated with cereal mildews, and fusariosis or smuts/spots [[1], [2], [3], [4]], and subsequently have a major impact on many populations globally, especially those who rely on cereals (like maize, wheat, and barley) as a staple food source (FAOSTAT, 2019). Many of the crop protection strategies and or plant disease management approaches used to eradicate or manage these crop invaders, relies mainly on the use of resistant crop cultivars and or synthetic antimicrobials [5]. More recently, however, the use of safer/environmentally friendly microbe-derived antimicrobials is becoming more popular [6,7]. Despite this, however, none of the strategies have led to the total eradication of these harmful phytopathogens, which regularly re-emerge and remain prevalent in many regions of the developing world [5,8]. The many precautionary measures put in place to prevent pre-harvest plant damage or post-harvest/storage crop loss (e.g. sun drying, air drying, and chemical application) by the unwanted toxicogenic microbes are considered insufficient, since regular accounts of both human and livestock poisoning from these toxins still exist [2,2,3,4,[9], [10], [11], [12]].
Reports about the histological, genomic, transcriptomic, and proteomic characteristics of these plant pathogens, the beneficial microbes, and the impact these have on the host plant are readily available [[13], [14], [15]]. The information about the real-time metabolic changes induced by the microbial activities (herein toxicogenic microbe-beneficial microbe) on the host plant and vice versa, is however scarce. Considering this, additional information on the metabolic changes induced during tripartite interactions (plant, toxicogenic microbes, and beneficial microbes), hereafter defined as plant-microbes symbiosis, interpreted alone or in combination with previous omics data, would significantly improve our understanding of the interaction. Understanding these interactions could ultimately lead to formulating new crop protection strategies for improving crop quality and yields [[16], [17], [18]]. Beneficial microbes herein are plant microbiota that improve the plant’s: (a) resistance to various plant stressors, (b) nutrient acquisition and growth capability, (c) defense response metabolism and or (d) produce anti-phytopathogenic biomolecules. Detailed reviews on beneficial microbes and their metabolic products functioning as plant growth promoters and/or phytopathogen biocontrollers (offering plant protection) are available for further reading [6,7,19,20]. Whilst early metabolic data characterizing various model plants (e.g. Arabidopsis thaliania and Medicago truncatula), phytopathogens (e.g. Aspergillus flavus, Pseudomonas syringae, and Botrytis cinerea) and plant beneficial-microbes (e.g. Bacillus thuringiensis, Penicillium chrysogenum, and Pseudomonas fluorescens) exist, the information was acquired using targeted research approaches. These studies identified/quantified specific phytopathogen virulence biomolecules [12,[21], [22], [23]], elucidated the mechanisms of action of growth-promoting or biocontrol compounds/organisms [[24], [25], [26], [27]], or studied a plant response to a specific phytopathogen/anti-phytopathogenic agent [[28], [29], [30], [31], [32], [33]].
More recently however, untargeted and semi-targeted metabolomic profiling, focused on understanding competitive, amensalistic, commensalistic, and mutualistic associations between plants, phytopathogens and beneficial microbes, are being utilized for these purposes [11,27,[34], [35], [36], [37]]. The metabolomic data on tripartite plant-microbes symbiosis is complex, yet limited, which necessitates this review. This paper focuses only on the bacterial and fungal investigations completed to date on this topic. The paper starts by explaining the various perturbations that can be associated with the aforementioned tripartite interactions, followed by a discussion of the literature describing how metabolomics has contributed to a better understanding of the changes to the phytopathogen metabolome during plant-microbe symbiosis, the metabolomics of beneficial microbes during concurrent host-phytopathogen interactions, and lastly how plant metabolome profiling could contribute to a better understanding of plant-microbes symbiosis.
2. Perturbations associated with a tripartite plant-microbes symbiosis
Studies investigating host plant-microbes interactions as either a one-way (amensalism and commensalism) or a two-way association (mutualism and competition) have been well-reported [38,39]. The application of metabolomics for efficient discrimination of metabolite origins and for understanding roles of all interacting organisms during tripartite plant-microbes symbiosis is, however, scarce. During the cohabitation of a phytopathogen, beneficial organism and the host plant, an alteration in each of their respective metabolomes is expected, due to the influx of foreign biomolecules from all the participants [40]. Specifically, the foreign metabolites originating from the interacting pathogen or beneficial organism, are expected to directly or indirectly alter various biosynthetic pathways of the host plant [17,21]. Invariably, during this tripartite coexistence, an influence (negative or positive) is exerted on the physiology of all the three participants in concert, since antibiosis brings about a disruption in the growth and proliferation of a pathogen, while the virulent pathogen causes a diseased state in the host plant. Considering this, characterizing the altered metabolic state of these interacting tripartite living systems (plant-microbes symbiosis) simultaneously, may assist to better understand the mechanisms and adaptations related to these interacting species.
Furthermore, it should be mentioned, that various teratogenic, carcinogenic, hepatotoxigenic, mutagenic, and neurotoxigenic substances (e.g. type B trichothecene, fumonisin, beauvericin, moniliformin, deoxynivalenol, fusaproliferin, patulin, and enniatins) produced by various mycotoxicogenic species (e.g. Aspergillus, Fusarium, and Penicillium sp.), are sometimes masked, or initially in an inactive state, and only become detectable when the plant is used as food for livestock, or during the processing of cereal foods for later human consumption. This subsequently makes these compounds and the changes induced by them during such circumstances difficult to assess or monitor in the infected pre-harvest and post-harvest plants. Herewith, metabolomics also offers a unique opportunity by which phytopathogen-specific metabolic biomarkers or plant disease biomarkers (indicative of infection or early plant disease biomarkers) might be identified in an untargeted manner, and then used to monitor the presence of the phytopathogen and/or the presence/progression of the disease.
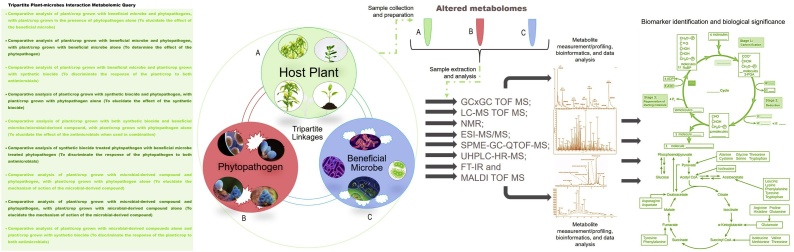
Studying in-field plant community dynamics presents a great challenge since free-growing plants interact with a multitude of microbes, and hence determining the exact cause of the perturbations/metabolite source, in a supposedly tripartite symbiosis, can become complicated. Hence an unbiased metabolite separation and or allocation method is required in many plant-microbe metabolomic investigations. Taking this into consideration, William Allwood, et al. [41], proposed a growth phase (time-course dependent) differential filtering and centrifugation dual metabolome profiling procedure, for the determination of metabolite source in more complex tripartite interaction studies, which might be used in free-growing plants. Here, the growth phases of the host plant and the interacting microbes are monitored independently, and the plants and microbes are harvested at different growth phases based on biomass monitoring. Fig. 1 illustrates the complex metabolomic profiles that we propose could be used for simultaneously investigating various tripartite plant-microbes symbiosis. This multi-comparison approach could aid in the understanding of metabolite sources and impacts during the different interactions.
Fig. 1.
Example of the complex multi-comparative metabolomic profiling perspectives for tripartite plant-microbe interactions. This comparison should facilitate metabolite source attribution. Key: Solid Phase Microextraction: (SPME); QTOF: Quadruple Time of Flight; GC: Gas Chromatography; LC: Liquid Chromatography; HR: High Resolution; MS: Mass spectrometry; UHP: Ultra-High Performance; NMR: Magnetic Resonance, FT-IR: Fourier Transform Infra-Red Spectroscopy, and MALDI TOF MS: matrix-assisted laser desorption/ionization, time-of-flight mass spectrometry analyzer.
Further considerations when planning a total metabolome profiling study (aimed at evaluating full metabolic signatures) investigating tripartite plant-microbes symbiosis are the strengths and limitations of each metabolomic analytical instrument, in the light of the expected outcomes. Various analytical instruments (e.g. headspace solid-phase microextraction gas chromatography (HS-SPME-GC)); ultrahigh-resolution liquid chromatography (UHLC-HR) coupled to time of flight or electrospray ionization mass spectrometry (TOF-MS/ESI-MS), selected by considering compound polarity and dimensionality, analytical sensitivity, resolution, repeatability, and reproducibility, are typically used for untargeted metabolomics. Concise reviews on metabolomic instrumentation, methodology and data analysis used, including the pros and cons of each analytical tool, are available for further reading [[42], [43], [44], [45]].
2.1. Mechanism of activity and roles of specific metabolites in tripartite plant-microbes interactions
Whilst the metabolic interplay within the tripartite plant-microbes linkage is expected to follow previously documented mechansims of plants defense, beneficial microbes mode of action, and phytopathogen virulence mechansism, new metabolite markers identified using untargeted metabolomics techniques, will allow for the identification of novel mechanisms related to the afore mentioned. The question, however, is that, can untargeted metabolomics provide an opportunity to discover novel mechanisms not related to previously document ones? If yes the researchers will thus avoid the inclination to screen for previously documented mechanisms. It is also important to note, however, that the impact of the mechanisms is dependent on several factors, not limited to the physiology of the host plant, the response of the phytopathogen to the plant’s defense mechanism, the phytopathogen’s virulence mechanism and the biocontrol mechanisms of the beneficial microbes investigated [46].
The exogenous and/or endogenous colonization of plant parts by toxicogenic or beneficial microbe result in the production and accumulation of numerous plant specific metabolites that play significant role in plant growth and health. For instance the colonization of host-plants by beneficial microbe led to the accumulation of various beneficial metabolites in the plant (e.g. catalpol, coumestrol, daidzein, gallic acid, myricitin, and tomatidine) which correspondingly led to: a. increased drought and salt tolerance, b. reduced pest damage, c. reduced pathogen proliferation, and d. higher nodulation in the host plant. [[47], [48], [49], [50]]. The reviews by Etalo, et al. [51] and Korenblum and Aharoni [47], provides information on plant metabolome distruptions induced by colonizing beneficial microbes and the significance of the specific metabolites produced due to the pertubations.
Concise reviews on the mechanisms underlining various plant metabolite’s (e.g. phytohormones, phytoalexins and defensins) role in plant disease resistance are available [[52], [53], [54], [55], [56], [57]]. For example, the mechanism of action of the antifungal defensin NaD1 from Nicotiana alata, includes membrane permeabilization of Fusarium hyphae, disruption of cell cytoplasm and subsequent induction of reactive oxygen species and cell death [54]. Furthemore, the two classical plant defense responses to phytopathogen attack (systemic acquired resistance (SAR) and induced systemic resistance (ISR)) have also been comprehensively discussed elswhere [[58], [59], [60]].
The virulence mechansims of phytopathogens (e.g. their host-plant debilitating mechanisms and antagonistic mechanisms) and the mechanism of action of beneficial microbes (direct and indirect biocontrol mechanisms) have also been adequately documented in existing reviews [46,[61], [62], [63], [64]]. For example, phytotoxins produced as virulent factors for some phytopathogens (e.g. P. syringae spp. and Rhizobium spp), function by distrupting the metabolism of the host-plant, via elevated ethylene production, which in turn reduces plant nodulation and plant biomass. Furthermore, various beneficial microbes have previously been reported to produce rhizobitoxine and 1-aminocyclopropane-1-carboxylic acid (ACC) deaminases, which inhibit the both synthesis and function of the ethylene precursor ACC and ACC synthase enzyme respectively [62,65,66] – a direct biocontrol mechanism. Some beneficial microbes also produce lytic metabolites (e.g. cellulases, chitinases, proteases, and -1,3 glucanases), capable of lysing the cell walls of many pathogenic fungi, while others produce siderophores (iron sequesters), preventing the acquisition of iron by such phytopathogens, subsequently inhibiting phytopathogen growth and colonization [62] – an indirect biocontrol mechanism.
The kind of microbial colonizer (phytopathogen or beneficial microbe) and the type of colonization (unilateral or multilateral), impacts the metabolite levels in the host-plant metabolome – and determines to a large extent the mechanisms that will be in play during the tripartite linkages. Often, unilateral or multilateral microbial colonization could lead to an accumulation of specific plant metabolite classes like the alkaloids, benzoxazinoids, isoprenoids, lignans, oxylipins, phenolics, and terpenes. In other situations, depending on the colonization type (unilateral or multilateral) and the colonizers (phytopathogen or beneficial microbe), the metabolite levels in the host-plant metabolome decreases or remains unchanged. According to Rodriguez, et al. [67], the host-plant colonization patterns of phytopathogens and beneficial microbes, and the subsequent metabolite changes may be identical, and the mechanisms by which the plant differentiates the origins of such, still needs to be understood. Untargeted metabolomics shows promise for elucidating such.
3. Phytopathogen metabolome changes during plant-microbes symbiosis
Recently, Azzollini, et al. [68], applied HS-SPME-GC–MS and LC-HRMS metabolomics to investigate the dynamics by which volatile and non-volatile metabolites are altered during fungal cohabitation. The study profiled the metabolome of two grapevine pathogens; Eutypa lata and Botryosphaeria obtusa (in co-culture), and investigated their individual responses to a commercially available 2-nonanone antifungal metabolite, the origin of which was not specified. Complete inhibition of the two grapevine pathogens was observed after seven days. The other major volatile and non-volatile metabolites identified in the study included decane, a yet unidentified compound from the sesquiterpene class and O-methylmellein.
A proton nuclear magnetic resonance (1H NMR) metabolomics approach was used by Sevastos, et al. [69], to characterize the metabolic disturbances induced in the metabolome of wild type F. graminearum, four carbendazim-resistant F. graminearum strains, (after treatment with the synthetic fungicide carbendazim) and an untreated F. graminearum strain. The authors observed a positive correlation among some metabolite levels (upregulated: l-serine, d-glucose, l-methionine, l-glutamate, l-phenylalanine, pyroglutamate, and citrate; down-regulated: threonine, d-myo-inositol, l-sucrose, and malate) detected in the wild and resistant F. graminearum strains, which could be later exploited in biomarker identification/selection for disease detection and monitoring. Prior to that, using LC—MS, Farrés, et al. [70] described the altered metabolome induced by toxic copper (Cu(II)) residues on a laboratory wine yeast strain, Saccharomyces cerevisiae (BY4741). Significant increases were recorded in the metabolic levels of trehalose, nicotinate d-ribonucleoside, l-glutamic acid, and nicotinamide d-ribonucleotide with a resultant decrease in the concentration of glutathione. Although the yeast's growth was not affected by sublethal concentrations of Cu(II), higher Cu(II) concentration led to its DNA damage and oxidative stress. Thus affirming the potential negative impact of Cu(II) containing fungicides on the wine yeasts (e.g. S. cerevisiae). High level copper residues in grapes may cause slow or stuck fermentation due to distruption of the beneficial S. cerevisiae metabolome. These studies summarized in Table 1 show the capacity of metabolomics to characterize the mechanisms associated with the effects of antimicrobial agents on the plant microflora.
Table 1.
Recent metabolomic elucidation of phytopathogen metabolome changes during plant-microbes symbiosis.
| Profiled metabolome: interacting phytopathogen | Interactions and study conditions | Significant metabolites identified | Metabolomic analytical resource | References |
|---|---|---|---|---|
| a. E. lata and B. obtuse; | a. Phytopathogen (E. lata and B. obtuse) + beneficial compound (2-nonanone; in vitro | a. 2-nonanone and O-methylmellein; | a. HS-SPME-GC-MS and LC-HRMS; | a. [68]; |
| b. Wild type F. graminearum and carbendazim-resistant F. graminearum and | b. Phytopathogen (wild type F. graminearum and carbendazim-resistant F. graminearum) + fungicide (carbendazim); in vitro | b. l-glutamate, pyroglutamate, l-methionine, l-phenylalanine, d-myo-inositol, and l-threonine; and | b. 1H NMR and | b. [69]; and |
| c. P. syringae pathovar tomato (Pst.) | c. Phytopathogen (P. syringae pathovar tomato) + A. thaliana; in vitro growth chamber experiment | c. Unspecified | c. FT-IR | c. [41] |
Key: HS-SPME: Headspace-Solid Phase Microextraction; GC: Gas Chromatography; LC: Liquid Chromatography; HR: High Resolution; MS: Mass spectrometry; 1H NMR: Nuclear Magnetic Resonance and FT-IR: Fourier Transform Infra-Red Spectroscopy.
4. Beneficial-microbe metabolome changes during plant-microbes symbiosis
Metabolomics of late has been used to characterize the altered metabolic state of various beneficial microbes that positively influence plant viability and growth, and those offering protection from invading phytopathogens (Table 2). An untargeted metabolomics investigation by Danquah, et al. [71], characterized the altered metabolic responses of marine-adapted fungal isolates to phytopathogenic bacteria (Ralstonia solanacearum and P. syringae), and a fungal (B. cinerea and Magnaporthe oryzae) challenge. From the mono- and co-culture studies performed for natural product discovery, several known (e.g. mitorubrins, 3,4-dihydromitorubrinol acetate, emerimicins, cytochalasins, ophiobolins, ustilaginoidins, eremophilanes, tenuazonic acid and sclerosporins) and novel metabolites (e.g. ergone) with proven antimicrobial bioactivity were characterized. The detected novel metabolites suggest the activation of cryptic biosynthetic pathways in the beneficial marine fungal isolates.
Table 2.
Recent applications of metabolomics in elucidating beneficial-microbe metabolome changes during plant-microbes symbiosis.
| Profiled metabolome: interacting beneficial microbe | Interactions and study conditions | Significant metabolites identified | Metabolomic analytical resource | References |
|---|---|---|---|---|
| a. Marine-adapted fungal isolates (Emericellopsis sp.; Hypoxylon sp.; Alternaria sp.; Acremonium sp., and Cosmospora sp.); | a. Beneficial marine-adapted fungal isolates + phytopathogen (P. syringae and R. solanacearum); in vitro; | a. Mitorubrinol, mitorubrinic acid, cytochalasins, emerimicins, cephalosporins, ophiobolins and zervamicin; | a. UPLC–QTOF–HRMS/MS; | a. [71]; |
| b. S. cerevisiae; | b. Beneficial microbe (S. cerevisiae) + xenobiotics (copper); in vitro; | b. Glutathione, l-Dihydroorotiacid and l-aspartic acid; | b. LC-MS; | b. [70] |
| c. Pseudomonads (P. fluorescens RA12, P. protegens Pf-5, P. putida KT2440, and P. putida S12); and | c. Beneficial microbe (Pseudomonads) + growth inhibitor (glyphosate); in vitro; and | c. Tryptophan, phenylalanine, shikimate-3-phosphate, citrulline, thymidine, fumarate, valine, glutamine, and ornithine; and | c. 13C-assisted LC-MS; and | c. [88]; and |
| d. Trichoderma asperellum | d. Beneficial microbe T. asperellum + phytopathogen (Phytophthora capsici); in vitro | d. Viridepyronone, virone, koninginin D, gliotoxin, and acetyltetrahydroxyanthraquinone | d. HPLC and LC-ESI-MS | d. [89] |
Key: UHP: Ultra-High Performance; QTOF: Quadruple Time of Flight; LC: Liquid Chromatography; HR: High Resolution; MS: Mass spectrometry; ESI: Electrospray ionization.
Of interest are the glyphosate-induced perturbations in some Pseudomonad’s metabolome reported by Aristilde et al. (2017). The metabolome disruptions of four beneficial Pseudomonas strains (P. fluorescens RA12, Pseudomonas protegens Pf-5, Pseudomonas putida KT2440, and P. putida S12), when grown under different concentrations of succinate and glyphosate (a toxic component of herbicides) were elucidated by 13C-assisted LC–MS metabolomics. Glyphosate negatively affected the growth of P. putida S12 and P. putida KT2440, while P. fluorescens RA12 and P. protegens were unperturbed in low or high glyphosate concentrations. The study confirmed that glyphosate targets the aromatic amino acid biosynthetic pathway of the affected organisms. Other major biomarkers identified were tryptophan, phenylalanine, shikimate-3-phosphate, citrulline, thymidine, fumarate, valine, glutamine, and ornithine. Their results suggest that the simultaneous use of glyphosate-containing herbicides might compromise the use of these beneficial microbes in an agricultural setting.
5. Plant metabolome changes during plant-microbes symbiosis
Metabolomic studies investigating various plant responses to either toxicogenic or beneficial microbe perturbations is far more prevalent in the literature compared to the two previously discussed sections (section 3 and 4) (Table 3). A study by Saia, et al. [72], describes one of the earliest attempts to understand tripartite plant-beneficial microbes symbiosis. Using GC-TOF-MS and HILIC-Q-TOFMS (hydrophilic interaction chromatography time-of-flight mass spectrometry), the symbiotic impact of arbuscular mycorrhizal fungi (AMF) and plant growth-promoting rhizobacteria (PGPR) consortium on the metabolome of durum wheat (Triticum durum Desf.) was determined. By comparing the wheat root metabolite profiles from singular AMF treatments, with those from co-culture AMF-PGPR, the authors showed that soil inoculation with AMF, either alone or in combination with PGPR, markedly increased wheat root colonization. They also compared the metabolome of nitrogen-deficient and phosphorus-rich wheat with the aforementioned treatment combinations. Xilitol was a major upregulated metabolite annotated in the study, and depending on the treatment combination, various other compounds including carnitines, d-arabitol, pipecolic acid, 2-oxoglutarate, pyruvate, zymosterol, choline group, ethanolamines, and l-arabinose were upregulated or downregulated. From the study, the authors concluded that only AMF impacted wheat metabolome reprogramming, while the PGPR had no significant additional effect.
Table 3.
Recent metabolomic elucidation of plant host metabolome changes during plant-microbes symbiosis.
| Profiled metabolome: plant host | Interactions and study conditions | Significant metabolites identified | Metabolomic analytical resource | References |
|---|---|---|---|---|
| a. Wheat; | a. Wheat+ beneficial microbes (AMF + PGPR); in planta field trial; | a. Xilitol, carnitines, d-arabitol, pipecolic acid, 2-oxoglutarate, pyruvate, zymosterol, choline group, ethanolamines, and l-arabinose; | a. GC-TOF-MS and HILIC-Q-TOF-MS; | a. [72]; |
| b. Grape berries; | b. Grape berries + phytopathogen (B. cinerea, Penicillium expansum, Aspergillus niger and A. carbonarius); in vitro microplate and zip-lock plastic bag assay; | b. 1,5-dimethylnaphthalene, unidentified sesquiterpenes, 2-(4-hexyl-2,5-dioxo-2,5-dihydrofuran-3-yl)acetic acid, m-cresol and γ-nonalactone; | b. SPME-GC-QTOF-MS; | b. [81]; |
| c. Barley, wheat and rice; | c. Barley, wheat and rice + pathogen cassette (Lr34 resistance gene); in planta hydroponic experiment; | c. Gentisic acid O-glucoside, C-glycosylated flavones, isoorientin-7-2″-di-O- glucoside and hordatines; | c. UHPLC-HR-MS; | c. [73]; |
| d. Potato leaf; | d. Potato leaf + Phosphite; in planta field trial; | d. Caffeic acid, salicylic acid, and chlorogenic acid; | d. GC-TOF-MS; | d. [90]; |
| e. Apple fruit; | e. Apple fruit + phytopathogen (P. expansum); in vitro growth chamber experiment; | e. Fructose, malic acid, shikimic acid, ascorbic acid and glutathione; | e. HPLC; | e. [75]; |
| f. A. thaliana cell; | f. A. thaliana + phytopathogen (P. syringae); in vitro growth chamber experiment; | f. Unspecified; | f. FT-IR; | f. [41]; |
| g. Soybean; | g. Soybean + phytopathogen (R. solani); in vitro growth chamber experiment | g. Coumarins, phytoalexins, and flavonoids; | g. GC-MS | g. [32]; |
| h. Tomato; | h. Tomato + phytopathogen (B. cinerea and P. syringae) + beneficial compound (hexanoic acid); in planta controlled experiment; | h. 1-methyltryptophan; | h. UHPLC-MS/GC-MS; | h. [37]; |
| i. Sugarcane bud setts; | i. Sugar cane bud setts + Sporisorium scitamineum; in planta (green-house); | i. Lyxose, glycerate, raffinose, and phenylpropanoid; | i. GC-TOF-MS and LC-ESI-MS/MS; | i. [74]; |
| j. Tomato; | j. Tomato + Trichoderma metabolites (6-pentyl-2H-pyran-2-one and harzianic acid); in vitro plant growth assay; | j. Alanine, arginine, asparagine, fructose galactose, glucose, glutamine, leucine, methionine, phenylalanine, sucrose, threonine, trigonelline, tyrosine, and valine; | j. HRMAS-NMR; and | j. [76]; |
| k. Citrus leaves; and | k. Citrus seeds + Candidatus Liberibacter asiaticus; in planta controlled experiment; and | k. Asparagine, choline, glucose, malic acid, maltose, proline, sucrose, threonine, trigonelline, quinic acid, and uridine; and | k. NMR; | k. [77]; and |
| l. Maize root | l. Wild type (WT) BX regulated maize (Zea mays cv.W22) root + BX deficient W22 mutant; in planta (green-house) | l. Dihydroxy-7-methoxy-1,4-benzoxazin-3-one and 2,4-dihydroxy-1,4-benzoxazin-3-one | l. UPLC-Q-TOF-MS | l. [78] |
Key: GC: Gas Chromatography; HRMAS: High-Resolution Magic-Angle-Spinning; HILIC: Hydrophilic interaction; QTOF: Quadruple Time of Flight; LC: Liquid Chromatography; HR: High Resolution; MS: Mass spectrometry; UHP: Ultra-High Performance; SPME: Solid Phase Microextraction; ESI: Electrospray ionization; and FT-IR: Fourier Transform Infra-Red Spectroscopy.
Using both GC–MS and LC–MS metabolomics, Bucher, et al. [73], elucidated the changes induced by Lr34 (a multi-phytopathogen resistance-conferring gene), in field-grown and transgenic green-house cultivated species of barley, wheat, and rice. The plants analysed were with or without rusts and powdery mildew disease. Although most of the secondary metabolites detected in the study were unidentifiable, nine primary metabolites and 16 lipids were disrupted in barley plants. Increased glucose and fructose levels were recorded in the barley plants, while down-regulation of dehydro-ascorbate was observed. Overall, 84 primary metabolites were identified from the extracts of the transgenic plants that included sugars, amino acid derivatives, polyols, organic acids, and several lipid classes. Using an Orbitrap MS (direct infusion) and GC–MS approach, Aliferis, et al. [32], also carried out a time-course monitoring of the response of soybeans to Rhizoctonia solani infection. The approach also involved constructing a comprehensive soybean metabolite library, which subsequently accelerated the process of metabolite identification and interpretation of data generated during the study. Biomolecules including coumarins, phytoalexins, and flavonoids, which enhanced soybean’s defense traits against biotic stress, were identified in the pool of metabolites detected. What is of additional value, apart from the mechanisms discoveries, is that this study approach can subsequently be adopted in other cereal grain investigations.
A similar GC-TOF-MS and LC-ESI-MS/MS combination approach was used by Schaker, et al. [74], to elucidate the metabolic changes of a seven-month-old (green-house grown) sugarcane bud setts, following artificial treatment with the fungal pathogen Sporisorium scitamineum. Quantitative alterations in a subset of 73 metabolites, from the plant metabolome with significant xylose, glycerate, and raffinose upregulation, were identified by GC-TOF-MS. These disruptions were determined to impact the cell wall precursors, amino acid and phenylpropanoid biosynthesis, and other major energy pathways. Furthermore, some rare antifungal-associated biomolecules were identified using the LC-ESI-MS. Most recently, Žebeljan, et al. [75], investigated the altered metabolome induced by Penicillum expansum on apples, with the most prominent changes to be in the ascorbate-glutathione pathway of the fruit, and a subsequent reduction of glutathione and shikimic acid levels as disease severity increased.
A high-resolution magic-angle-spinning nuclear magnetic resonance (HRMAS-NMR) spectroscopy was employed by [76], to determine the metabolome changes in tomato leaves (Solanum lycopersicum) due to treatments of tomato seeds with two Trichoderma biocontrol metabolites (6-pentyl-2H-pyran-2-one and harzianic acid). Data generated revealed that the tomato leaves metabolome, its seedling fresh weight and seed germination rates were dependent upon the treatment doses of the Trichoderma metabolites. Altogether, γ-aminobutyric acid (GABA) and acetylcholine levels in both treatments showed a remarkable increase relative to the control samples. Metabolites with upward modulation in 6-pentyl-2H-pyran-2-one treated samples included arginine, glutamine, leucine, methionine, phenylalanine, sucrose, threonine, trigonelline, tyrosine, and valine, while those with upward modulation in the harzianic acid treatment were alanine, asparagine, galactose, phenylalanine and sucrose. Also relative to the controls the major metabolites with reduced concentration were glucose and fructose. The study corroborated previous claims of the biocontrol properties of Trichoderma metabolites.
Another NMR analysis by Padhi, et al. [77], elucidated the root metabolome response of two citrus plant varieties (‘Lisbon’ lemon (Citrus limon L. Burm, f.) and ‘Washington Navel’ orange (Citrus sinensis (L.) Osbeck) to Candidatus Liberibacter asiaticus (CLas) infection. CLas is the causative agent of the citrus greening disease - Huanglongbing (HLB). In general, a significant difference in 27 water-soluble metabolites where reported (asparagine, choline, glucose, malic acid, maltose, proline, sucrose, threonine, trigonelline, quinic acid, and uridine), and all of which were associated with plant defense mechanisms. More specific observations worth mentioning was a significant decrease in the levels of quinic acid and malic acid in the lemon roots only, and some metabolite response overlap in the responses of the two plants to CLas. The latter included elevated trigonelline levels, reduced levels of all the sugar metabolites measured, and reduced concentrations of choline, uridine, asparagine, and proline in both species. This study subsequently showed that the management of citrus greening disease (Huanglongbing) might require a varietal treatment or control approach.
The role of the tryptophan-derived heteroaromatic metabolites; benzoxazinoids (BXs), as regulators of plant-microbes interaction, are now becoming better understood. As part of a larger study to correlate the effects of Bx-regulated root metabolites with Bx-dependent rhizosphere microbiota, Cotton, et al. [78] determined, using an untargeted UPLC-Q-TOF-MS metabolomics analysis, the impact of the BXs metabolites on the maize root metabolome. In the study, a comparison was made between the metabolome of wild type (WT) BX regulated maize (Zea mays cv.W22) root and a BX deficient W22 mutant (the mutation brought about by inserting transposons at 3 different steps (bx1, bx2 and bx6) of the BX biosynthesis pathway). Data from the analysed crown and primary maize roots extracts indicated that the bx mutations did not significantly affect the growth and development of the maize mutants. Although the levels of the BXs (dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA) and 2,4-dihydroxy-1,4-benzoxazin-3-one (DIBOA)) in both the crown and primary roots of the bx1 and bx2 mutants decreased dramatically in comparison with that in the WT, the roots of bx6 mutants, however, showed elevated levels of DIBOA but reduced levels of DIMBOA in comparison with the WT. Whilst the bx1 and bx2 mutations had similar impact on the total BX production, the bx6 mutation, however, had a relatively minor effect on the root BX composition. Overall, the WT and bx roots profiling indicated that the bx1 and bx2 mutations significantly impact the root metabolome and corroborated previous reports of BXs role in metabolic regulation and differentiation of maize roots [78]; and that is, that BXs are involved in the regulation of a vast group of secondary root metabolites [[78], [79], [80]].
In another effort to determine the changes to organoleptic properties of fungal rot grapes, Schueuermann, et al. [81], exposed grape berries to P. expansum, B. cinerea, A. niger or Aspergillus carbonarius, and subsequently used an untargeted GC–MS metabolomics approach to identify and quantify the altered volatile metabolites profiles because of the resulting rot. The authors characterized B. cinerea incursion to result in the production of 1,5-dimethylnaphthalene and some various sesquiterpenes; A. niger (2-(4-hexyl-2,5-dioxo-2,5-dihydrofuran-3-yl) acetic acid), A. carbonarius (phenylethyl alcohol and β -damascenone), and P. expansum (m-cresol and γ-nonalactone). The study subsequently suggests that the biomarkers can discriminate causative agents of grape bunch rot and can be used in distinguishing between crop phytopathogens causing similar diseases.
Apart from those phytopathogens causing similar plant diseases, efficiently monitoring plant defense responses or susceptibility to pathogenic attack in unrelated phytopathogens (having different life cycles and virulence mechanisms) is also necessary. This should also be considered for beneficial microbes with dissimilar growth patterns and biocontrol mechanisms. Furthermore, in order for a negative or positive symbiosis to occur in a host plant, the phytopathogen or beneficial microbe must efficiently colonize a plant and each tripartite participant (negative colonizer, positive colonizer, and host plant) subsequently elicit an appropriate metabolic response [82]. Considering this, various studies have been performed identifying significant phytopathogen virulence-related metabolites and elucidating their roles and mechanisms in plant pathogenesis [35,83]. The study by Camañes, et al. [37], describes the complexity of assessing the metabolic perturbations of tripartite plant-microbes symbiosis. Using an untargeted UHLC/MS and GC–MS global metabolomics approach, the researchers compared the metabolomics profiles of infected tomato plants, primed tomato plants, and primed plus infected tomato plants. As hypothesized, the metabolic profiles captured from the metabolome of the phytopathogen-infected tomato were different. This correlated with the distinct lifecycle and virulence mechanisms of the phytopathogens investigated. Additionally, 1-methyltryptophan was identified as a unique biomarker associated with the metabolome of the phytopathogen-infected tomato and the hexanoic acid primed tomato metabolome. The study revealed that metabolites elicited by plants in response to biotic and or abiotic perturbations are source dependent.
6. Biomarker application in crop enhancement and protection strategies
Metabolomics offers a unique approach by which phytopathogen-specific metabolic biomarkers or plant disease/defense biomarkers, could be identified using an untargeted approach and then used to monitor phytopathogen infectivity and or disease progression. Furthermore, functional biomolecules associated with the biocontrol organisms, can be isolated or synthesized and used in phytopathogen control, or to evoke anti-phytopathogenic mechanisms in plants. For example, specific biomarkers and or biochemical processes identified during maize-Fusarium graminearum-Bacillus amyloliquefaciens or soybean-Rhizoctonia- B. amyloliquefaciens interaction, would most likely contribute to a better understanding of the metabolic regulation of all the interacting living systems, providing valuable insights potentially useful in plant breeding, metabolic bioengineering, and agrochemistry research. Bio-products like resistant crop cultivars, robust secondary metabolite-producing beneficial microbes, and biofungicides can then be cultivated/cultured/produced and used in planta.
Another study by Khan, et al. [84], identified anti-drought stress biomarkers (malonate, leucine, 5-oxo-l-proline, saccharic acid, trans-cinnamate, succinate, and glyceric acid) in the chickpea (Cicer arietinum L.) metabolome, when treated with plant growth regulators (salicylic acid and putrescine) and PGPR consortium (B. thuringiensis, Bacillus subtilis, and Bacillus megaterium). Deliberate metabolic reprogramming of the chickpeas targeting the biomarker synthesizing pathways subsequently resulted in drought-tolerant chickpea varieties. Considering the above investigations, metabolomics could also be used to identify various biomarkers in rhizospheric and bulk soil, to assess/monitor plant infection pre-harvest and crop invasion post-harvest, for the purpose of reducing the exposure of livestock and humans to contaminated grains and or farmlands.
7. Concluding remarks
The application of untargeted metabolomics in the fields of pharmacology, chemistry, and clinical medicine, towards the discovery of molecular networks and metabolite interactions/elicitations, biomolecule structures, disease diagnostics, and biomarker identification, is already well established. The application of untargeted metabolomics towards improved crop/plant protection and food security strategies, however, is still relatively new, and the work done to date shows excellent scope, especially for improving our understanding of the overall adaptive physiology of the plant, and that of the interacting microbes [[85], [86], [87]]. A better understanding of the intra- and inter-species microbial interactions occurring at different heterogeneous levels within the plant habitat is imperative. Furthermore, identifying the systemic responses of various crops, to pathogenic stress, and the biocontrol thereof, would enable the crop scientist to identify unique metabolic markers that can be applied toward the early detection of a phytopathogen or its metabolites, in asymptomatic crops [15], as well as towards the development of biofungicides for example, for use during pre-harvest, post-harvest and large scale storage of crops. Novel insights into phytopathogen metabolism using metabolomics would also lead to a better understanding of phytopathogen colonization and pesticide tolerance.
Finally, although tripartite plant-microbes symbiosis metabolomics could be complicated due to the diversity of the associated biomolecules and large data generated, recent improvements to metabolomics methodology (semi-targeted and untargeted), equipment, and chemometrics/bioinformatics have led to faster, easier and more repeatable data acquisition. We foresee an exponentially increased identification and application of metabolite biomarkers in controlled and semi-controlled planting systems in the near future, and if properly integrated into crop protection strategies, food insecurity, and many other challenges farmers face in disease prevalent regions of the globe could be mitigated.
Funding
No specific funding was received for the study.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a conflict of interest.
Acknowledgment
First author appreciates North-West University for Post-Doctoral fellowship.
References
- 1.Hanvi D.M., Lawson-Evi P., De Boevre M., Goto C., De Saeger S., Eklu-Gadegbeku K. Natural occurrence of mycotoxins in maize and sorghum in Togo. Mycotoxin Res. 2019:1–7. doi: 10.1007/s12550-019-00351-1. [DOI] [PubMed] [Google Scholar]
- 2.Franco L.T., Petta T., Rottinghaus G.E., Bordin K., Gomes G.A., Oliveira C.A.F. Co-occurrence of mycotoxins in maize food and maize-based feed from small-scale farms in Brazil: a pilot study. Mycotoxin Res. 2019;35:65–73. doi: 10.1007/s12550-018-0331-4. [DOI] [PubMed] [Google Scholar]
- 3.Chilaka C.A., De Boevre M., Atanda O.O., De Saeger S. Occurrence of Fusarium mycotoxins in cereal crops and processed products (Ogi) from Nigeria. Toxins. 2016;8:342. doi: 10.3390/toxins8110342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H.J., Ryu D. Worldwide occurrence of mycotoxins in cereals and cereal-derived food products: public health perspectives of their co-occurrence. J. Agric. Food Chem. 2017;65:7034–7051. doi: 10.1021/acs.jafc.6b04847. [DOI] [PubMed] [Google Scholar]
- 5.Abdallah M.F., De Boevre M., Audenaert K., Haesaert G., De Saeger S. Highlight report: mycotoxins as food contaminants in Africa—challenges and perspectives. Arch. Toxicol. 2018;92:2151–2152. doi: 10.1007/s00204-018-2203-2. [DOI] [PubMed] [Google Scholar]
- 6.Ghorbanpour M., Omidvari M., Abbaszadeh-Dahaji P., Omidvar R., Kariman K. Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol. Control. 2018;117:147–157. [Google Scholar]
- 7.Rosier A., Medeiros F.H.V., Bais H.P. Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil. 2018;428:35–55. [Google Scholar]
- 8.Kagot V., Okoth S., De Boevre M., De Saeger S. Biocontrol of Aspergillus and Fusarium mycotoxins in Africa: benefits and limitations. Toxins. 2019;11:109. doi: 10.3390/toxins11020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nafuka S.N., Misihairabgwi J.M., Bock R., Ishola A., Sulyok M., Krska R. Variation of fungal metabolites in sorghum malts used to prepare Namibian traditional fermented beverages Omalodu and Otombo. Toxins. 2019;11:165. doi: 10.3390/toxins11030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanvi D.M., Lawson-Evi P., De Boevre M., Goto C.E., De Saeger S., Eklu-Gadegbeku K. Natural occurrence of mycotoxins in maize and sorghum in Togo. Mycotoxin Res. 2019 doi: 10.1007/s12550-019-00351-1. [DOI] [PubMed] [Google Scholar]
- 11.Kim D.-H., Hong S.-Y., Kang J.W., Cho S.M., Lee K.R., An T.K., Lee C., Chung S.H. Simultaneous determination of multi-mycotoxins in cereal grains collected from South Korea by LC/MS/MS. Toxins. 2017;9:106. doi: 10.3390/toxins9030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juan C., Ritieni A., Mañes J. Occurrence of Fusarium mycotoxins in Italian cereal and cereal products from organic farming. Food Chem. 2013;141:1747–1755. doi: 10.1016/j.foodchem.2013.04.061. [DOI] [PubMed] [Google Scholar]
- 13.González-Fernández R., Valero-Galván J., Gómez-Gálvez F.J., Jorrín-Novo J.V. Unraveling the in vitro secretome of the phytopathogen Botrytis cinerea to understand the interaction with its hosts. Front. Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gauthier L., Atanasova-Penichon V., Chéreau S., Richard-Forget F. Metabolomics to decipher the chemical defense of cereals against Fusarium graminearum and deoxynivalenol accumulation. Int. J. Mol. Sci. 2015;16:24839. doi: 10.3390/ijms161024839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galeano Garcia P., Neves dos Santos F., Zanotta S., Eberlin M.N., Carazzone C. Metabolomics of Solanum lycopersicum infected with Phytophthora infestans leads to early detection of late blight in asymptomatic plants. Molecules. 2018;23:3330. doi: 10.3390/molecules23123330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hautbergue T., Jamin E., Debrauwer L., Puel O., Oswald I. From genomics to metabolomics, moving toward an integrated strategy for the discovery of fungal secondary metabolites. Nat. Prod. Rep. 2018;35:147–173. doi: 10.1039/c7np00032d. [DOI] [PubMed] [Google Scholar]
- 17.Adeniji A.A., Loots D.T., Babalola O.O. Bacillus velezensis: phylogeny, useful applications, and avenues for exploitation. Appl. Microbiol. Biotechnol. 2019;103:3669–3682. doi: 10.1007/s00253-019-09710-5. [DOI] [PubMed] [Google Scholar]
- 18.Romero F.M., Marina M., Pieckenstain F.L., Rossi F.R., Gonzalez M.E., Vignatti P., Gárriz A. Metabolic Engineering for Bioactive Compounds, Springer; 2017. Gaining Insight Into Plant Responses to Beneficial and Pathogenic Microorganisms Using Metabolomic and Transcriptomic Approaches; pp. 113–140. [Google Scholar]
- 19.Mendes R., Garbeva P., Raaijmakers J.M. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 20.Babalola O.O. Beneficial bacteria of agricultural importance. Biotechnol. Lett. 2010;32:1559–1570. doi: 10.1007/s10529-010-0347-0. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y.-J., Wang C., Ho W.E., Ong C.N. Recent developments and applications of metabolomics in microbiological investigations. Trac Trends Anal. Chem. 2014;56:37–48. [Google Scholar]
- 22.Shank R.A., Foroud N.A., Hazendonk P., Eudes F., Blackwell B.A. Current and future experimental strategies for structural analysis of trichothecene mycotoxins—a prospectus. Toxins. 2011;3:1518–1553. doi: 10.3390/toxins3121518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., Zhou X., Naman C.B., Lu Y., Ding L., He S. Preparative separation and purification of trichothecene mycotoxins from the marine fungus Fusarium sp. LS68 by high-speed countercurrent chromatography in stepwise elution mode. Mar. Drugs. 2018;16:73. doi: 10.3390/md16020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuperlovic-Culf M., Rajagopalan N., Tulpan D., Loewen M.C. Metabolomics and cheminformatics analysis of antifungal function of plant metabolites. Metabolites. 2016;6:31. doi: 10.3390/metabo6040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trivedi D.K., Hollywood K.A., Goodacre R. Metabolomics for the masses: the future of metabolomics in a personalized world. New Horiz. Transl. Med. 2017;3:294–305. doi: 10.1016/j.nhtm.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adeniji A.A., Aremu O.S., Babalola O.O. Selecting lipopeptide‐producing, Fusarium‐suppressing Bacillus spp.: metabolomic and genomic probing of Bacillus velezensis NWUMFkBS10. 5. MicrobiologyOpen. 2018:e742. doi: 10.1002/mbo3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palyzová A., Svobodová K., Sokolová L., Novák J., Novotný Č. Metabolic profiling of Fusarium oxysporumf. Sp. Conglutinans race 2 in dual cultures with biocontrol agents Bacillus amyloliquefaciens, Pseudomonas aeruginosa, and Trichoderma harzianum. Folia Microbiol. (Praha) 2019 doi: 10.1007/s12223-019-00690-7. [DOI] [PubMed] [Google Scholar]
- 28.Bender C.L., Alarcón-Chaidez F., Gross D.C. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D., Fu J., Zhou R., Li Z., Xie Y., Liu X., Han Y. Formation of sclerotia in Sclerotinia ginseng and composition of the sclerotial exudate. PeerJ. 2018;6 doi: 10.7717/peerj.6009. e6009-e6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong X., Ling N., Wang M., Shen Q., Guo S. Fusaric acid is a crucial factor in the disturbance of leaf water imbalance in Fusarium-infected banana plants. Plant Physiol. Biochem. 2012;60:171–179. doi: 10.1016/j.plaphy.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Kleigrewe K., Aydin F., Hogrefe K., Piecuch P., Bergander K., Würthwein E.-U., Humpf H.-U. Structure elucidation of new fusarins revealing insights in the rearrangement mechanisms of the Fusarium mycotoxin fusarin C. J. Agric. Food Chem. 2012;60:5497–5505. doi: 10.1021/jf3009469. [DOI] [PubMed] [Google Scholar]
- 32.Aliferis K.A., Faubert D., Jabaji S. A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS One. 2014;9 doi: 10.1371/journal.pone.0111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nokhrina K., Ray H., Bock C., Georges F. Metabolomic shifts in Brassica napus lines with enhanced BnPLC2 expression impact their response to low temperature stress and plant pathogens. GM Crops Food. 2014;5:120–131. doi: 10.4161/gmcr.28942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Combès A., Ndoye I., Bance C., Bruzaud J., Djediat C., Dupont J., Nay B., Prado S. Chemical communication between the endophytic fungus Paraconiothyrium variabile and the phytopathogen Fusarium oxysporum. PLoS One. 2012;7 doi: 10.1371/journal.pone.0047313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu W., Pan X., Li F., Dong W. UPLC-QTOF-MS metabolomics analysis revealed the contributions of metabolites to the pathogenesis of Rhizoctonia solani strain AG-1-IA. PLoS One. 2018;13 doi: 10.1371/journal.pone.0192486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alsharif A.M.A., Choo Y.-M., Tan G.-H. Detection of five mycotoxins in different food matrices in the Malaysian market by using validated liquid chromatography electrospray ionization triple quadrupole mass spectrometry. Toxins. 2019;11:196. doi: 10.3390/toxins11040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camañes G., Scalschi L., Vicedo B., González-Bosch C., García-Agustín P. An untargeted global metabolomic analysis reveals the biochemical changes underlying basal resistance and priming in Solanum lycopersicum, and identifies 1-methyltryptophan as a metabolite involved in plant responses to Botrytis cinerea and Pseudomonas syringae. Plant J. 2015;84:125–139. doi: 10.1111/tpj.12964. [DOI] [PubMed] [Google Scholar]
- 38.Lugtenberg B. Springer; 2016. Principles of Plant-microbe Interactions. [Google Scholar]
- 39.Rey T., Schornack S. Interactions of beneficial and detrimental root-colonizing filamentous microbes with plant hosts. Genome Biol. 2013;14:121. doi: 10.1186/gb-2013-14-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feussner I., Polle A. What the transcriptome does not tell — proteomics and metabolomics are closer to the plants’ patho-phenotype. Curr. Opin. Plant Biol. 2015;26:26–31. doi: 10.1016/j.pbi.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 41.William Allwood J., Clarke A., Goodacre R., Mur L.A.J. Dual metabolomics: a novel approach to understanding plant–pathogen interactions. Phytochemistry. 2010;71:590–597. doi: 10.1016/j.phytochem.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Rempe C.S., Burris K.P., Lenaghan S.C., Stewart C.N. The potential of systems biology to discover antibacterial mechanisms of plant phenolics. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan M., Xu G. Current and future perspectives of functional metabolomics in disease studies–A review. Anal. Chim. Acta. 2018;1037:41–54. doi: 10.1016/j.aca.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 44.Dudzik D., Barbas-Bernardos C., García A., Barbas C. Quality assurance procedures for mass spectrometry untargeted metabolomics. A review, Journal of Pharmaceutical and Biomedical Analysis. 2018;147:149–173. doi: 10.1016/j.jpba.2017.07.044. [DOI] [PubMed] [Google Scholar]
- 45.Patejko M., Jacyna J., Markuszewski M.J. Sample preparation procedures utilized in microbial metabolomics: an overview. J. Chromatogr. B. 2017;1043:150–157. doi: 10.1016/j.jchromb.2016.09.029. [DOI] [PubMed] [Google Scholar]
- 46.Köhl J., Kolnaar R., Ravensberg W.J. Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 2019;10:845. doi: 10.3389/fpls.2019.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korenblum E., Aharoni A. Phytobiome metabolism: beneficial soil microbes steer crop plants’ secondary metabolism. Pest Manag. Sci. 2019 doi: 10.1002/ps.5440. [DOI] [PubMed] [Google Scholar]
- 48.Algar E., Gutierrez-Mañero F.J., Garcia-Villaraco A., García-Seco D., Lucas J.A., Ramos-Solano B. The role of isoflavone metabolism in plant protection depends on the rhizobacterial MAMP that triggers systemic resistance against Xanthomonas axonopodis pv. Glycines in Glycine max (L.) Merr. Cv. Osumi, Plant Physiology and Biochemistry. 2014;82:9–16. doi: 10.1016/j.plaphy.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Jain A., Singh A., Singh S., Singh H.B. Phenols enhancement effect of microbial consortium in pea plants restrains Sclerotinia sclerotiorum. Biol. Control. 2015;89:23–32. [Google Scholar]
- 50.Rivero J., Alvarez D., Flors V., Azcon-Aguilar C., Pozo M.J. Root metabolic plasticity underlies functional diversity in mycorrhiza-enhanced stress tolerance in tomato. New Phytol. 2018;220:1322–1336. doi: 10.1111/nph.15295. [DOI] [PubMed] [Google Scholar]
- 51.Etalo D.W., Jeon J.-S., Raaijmakers J.M. Modulation of plant chemistry by beneficial root microbiota. Nat. Prod. Rep. 2018;35:398–409. doi: 10.1039/c7np00057j. [DOI] [PubMed] [Google Scholar]
- 52.Hiruma K. Roles of plant-derived secondary metabolites during interactions with pathogenic and beneficial microbes under conditions of environmental stress. Microorganisms. 2019;7:362. doi: 10.3390/microorganisms7090362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piasecka A., Jedrzejczak‐Rey N., Bednarek P. Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 2015;206:948–964. doi: 10.1111/nph.13325. [DOI] [PubMed] [Google Scholar]
- 54.Khan R.S., Iqbal A., Malak R., Shehryar K., Attia S., Ahmed T., Khan M.A., Arif M., Mii M. Plant defensins: types, mechanism of action and prospects of genetic engineering for enhanced disease resistance in plants. 3 Biotech. 2019;9:192. doi: 10.1007/s13205-019-1725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bedini A., Mercy L., Schneider C., Franken P., Lucic-Mercy E. Unravelling the initial plant hormone signalling, metabolic mechanisms and plant defense triggering the endomycorrhizal symbiosis behavior. Front. Plant Sci. 2018;9:1800. doi: 10.3389/fpls.2018.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parisi K., Shafee T.M., Quimbar P., van der Weerden N.L., Bleackley M.R., Anderson M.A. Seminars in Cell & Developmental Biology. Elsevier; 2018. The evolution, function and mechanisms of action for plant defensins. [DOI] [PubMed] [Google Scholar]
- 57.Vriens K., Cammue B., Thevissen K. Antifungal plant defensins: mechanisms of action and production. Molecules. 2014;19:12280–12303. doi: 10.3390/molecules190812280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yi H.-S., Yang J.W., Ryu C.-M. ISR meets SAR outside: additive action of the endophyte Bacillus pumilus INR7 and the chemical inducer, benzothiadiazole, on induced resistance against bacterial spot in field-grown pepper. Front. Plant Sci. 2013;4 doi: 10.3389/fpls.2013.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vallad G., Goodman R. Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Science - CROP SCI. 2004;44 [Google Scholar]
- 60.Martinez-Medina A., Fernandez I., Sánchez-Guzmán M., Jung S., Pascual J., Pozo M. Deciphering the hormonal signalling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front. Plant Sci. 2013;4 doi: 10.3389/fpls.2013.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olanrewaju O.S., Glick B.R., Babalola O.O. Mechanisms of action of plant growth promoting bacteria, World J. Microbiol. Biotechnol. 2017;33:197. doi: 10.1007/s11274-017-2364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glick B.R. Plant growth-promoting bacteria: mechanisms and applications. Scientifica. 2012;2012 doi: 10.6064/2012/963401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pusztahelyi T., Holb I.J., Pócsi I. Plant-fungal interactions: special secondary metabolites of the biotrophic, necrotrophic, and other specific interactions. Fungal Metabolites. 2016:1–58. [Google Scholar]
- 64.Ahemad M., Kibret M. Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. Journal of King Saud University-Science. 2014;26:1–20. [Google Scholar]
- 65.Ishiga Y. Studies on mode of action of phytotoxin coronatine produced by Pseudomonas syringae pv. Tomato. J. Gen. Plant Pathol. 2017;83:424–426. [Google Scholar]
- 66.Xin X.-F., He S.Y. Pseudomonas syringae pv. Tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 2013;51:473–498. doi: 10.1146/annurev-phyto-082712-102321. [DOI] [PubMed] [Google Scholar]
- 67.Rodriguez P.A., Rothballer M., Chowdhury S.P., Nussbaumer T., Gutjahr C., Falter-Braun P. Systems biology of plant microbiome interactions. Mol. Plant. 2019 doi: 10.1016/j.molp.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 68.Azzollini A., Boggia L., Boccard J., Sgorbini B., Lecoultre N., Allard P.-M., Rubiolo P., Rudaz S., Gindro K., Bicchi C., Wolfender J.-L. Dynamics of metabolite induction in fungal co-cultures by metabolomics at both volatile and non-volatile levels. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sevastos A., Kalampokis I.F., Panagiotopoulou A., Pelecanou M., Aliferis K.A. Implication of Fusarium graminearum primary metabolism in its resistance to benzimidazole fungicides as revealed by 1H NMR metabolomics. Pestic. Biochem. Physiol. 2018;148:50–61. doi: 10.1016/j.pestbp.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 70.Farrés M., Piña B., Tauler R. LC-MS based metabolomics and chemometrics study of the toxic effects of copper on Saccharomyces cerevisiae. Metallomics. 2016;8:790–798. doi: 10.1039/c6mt00021e. [DOI] [PubMed] [Google Scholar]
- 71.Danquah C.A., Kakagianni E., Khondkar P., Maitra A., Rahman M., Evangelopoulos D., McHugh T.D., Stapleton P., Malkinson J., Bhakta S. Analogues of disulfides from Allium stipitatum demonstrate potent anti-tubercular activities through drug efflux pump and biofilm inhibition. Sci. Rep. 2018;8:1150. doi: 10.1038/s41598-017-18948-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saia S., Ruisi P., Fileccia V., Di Miceli G., Amato G., Martinelli F. Metabolomics suggests that soil inoculation with arbuscular mycorrhizal fungi decreased free amino acid content in roots of durum wheat grown under P-limited, P-rich field conditions. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bucher R., Veyel D., Willmitzer L., Krattinger S., Keller B., Bigler L. Combined GC- and UHPLC-HR-MS based metabolomics to analyze durable anti-fungal resistance processes in cereals. Chim. Int. J. Chem. 2017;71:156–159. doi: 10.2533/chimia.2017.156. [DOI] [PubMed] [Google Scholar]
- 74.Schaker P.D., Peters L.P., Cataldi T.R., Labate C.A., Caldana C., Monteiro-Vitorello C.B. Metabolome dynamics of smutted sugarcane reveals mechanisms involved in disease progression and whip emission. Front. Plant Sci. 2017;8:882. doi: 10.3389/fpls.2017.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Žebeljan A., Vico I., Duduk N., Žiberna B., Urbanek Krajnc A. Dynamic changes in common metabolites and antioxidants during Penicillium expansum-apple fruit interactions. Physiol. Mol. Plant Pathol. 2019;106:166–174. [Google Scholar]
- 76.Mazzei P., Vinale F., Woo S.L., Pascale A., Lorito M., Piccolo A. Metabolomics by proton high-resolution magic-angle-spinning nuclear magnetic resonance of tomato plants treated with two secondary metabolites isolated from Trichoderma. J. Agric. Food Chem. 2016;64:3538–3545. doi: 10.1021/acs.jafc.6b00801. [DOI] [PubMed] [Google Scholar]
- 77.Padhi E.M., Maharaj N., Lin S.-Y., Mishchuk D.O., Chin E., Godfrey K., Foster E., Polek M., Leveau J.H., Slupsky C.M. Metabolome and microbiome signatures in the roots of Citrus Affected by huanglongbing. Phytopathology. 2019;109:2022–2032. doi: 10.1094/PHYTO-03-19-0103-R. [DOI] [PubMed] [Google Scholar]
- 78.Cotton T.A., Pétriacq P., Cameron D.D., Al Meselmani M., Schwarzenbacher R., Rolfe S.A., Ton J. Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 2019:1. doi: 10.1038/s41396-019-0375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahmad S., Veyrat N., Gordon-Weeks R., Zhang Y., Martin J., Smart L., Glauser G., Erb M., Flors V., Frey M. Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol. 2011;157:317–327. doi: 10.1104/pp.111.180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meihls L.N., Handrick V., Glauser G., Barbier H., Kaur H., Haribal M.M., Lipka A.E., Gershenzon J., Buckler E.S., Erb M. Natural variation in maize aphid resistance is associated with 2, 4-dihydroxy-7-methoxy-1, 4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell. 2013;25:2341–2355. doi: 10.1105/tpc.113.112409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schueuermann C., Steel C.C., Blackman J.W., Clark A.C., Schwarz L.J., Moraga J., Collado I.G., Schmidtke L.M. A GC–MS untargeted metabolomics approach for the classification of chemical differences in grape juices based on fungal pathogen. Food Chem. 2019;270:375–384. doi: 10.1016/j.foodchem.2018.07.057. [DOI] [PubMed] [Google Scholar]
- 82.Pfeilmeier S., Caly D.L., Malone J.G. Bacterial pathogenesis of plants: future challenges from a microbial perspective. Mol. Plant Pathol. 2016;17:1298–1313. doi: 10.1111/mpp.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gai Y.-P., Han X.-J., LI Y.-Q., Yuan C.-Z., Mo Y.-Y., Guo F.-Y., Liu Q.-X., Ji X.-L. Metabolomic analysis reveals the potential metabolites and pathogenesis involved in mulberry yellow dwarf disease, Plant. Cell & Environment. 2014;37:1474–1490. doi: 10.1111/pce.12255. [DOI] [PubMed] [Google Scholar]
- 84.Khan N., Bano A., Babar M.D.A. Metabolic and physiological changes induced by plant growth regulators and plant growth promoting rhizobacteria and their impact on drought tolerance in Cicer arietinum L. PLoS One. 2019;14 doi: 10.1371/journal.pone.0213040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mhlongo M.I., Piater L.A., Madala N.E., Labuschagne N., Dubery I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018;9 doi: 10.3389/fpls.2018.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tenenboim H., Brotman Y. Omic relief for the biotically stressed: metabolomics of plant biotic interactions. Trends Plant Sci. 2016;21:781–791. doi: 10.1016/j.tplants.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 87.Tugizimana F., Mhlongo M.I., Piater L.A., Dubery I.A. Metabolomics in plant priming research: The way forward? Int. J. Mol. Sci. 2018;19:1759. doi: 10.3390/ijms19061759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aristilde L., Reed M.L., Wilkes R.A., Youngster T., Kukurugya M.A., Katz V., Sasaki C.R.S. Glyphosate-induced specific and widespread perturbations in the metabolome of soil Pseudomonas species. Front. Environ. Sci. 2017;5 [Google Scholar]
- 89.De la Cruz-Quiroz R., Ascacio-Valdés J.A., Rodríguez-Herrera R., Roussos S., Aguilar C.N. Phytopathogen biomass as inducer of antifungal compounds by Trichoderma asperellum under solid-state fermentation. In: Singh H.B., Keswani C., Reddy M.S., Sansinenea E., García-Estrada C., editors. Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms: Discovery and Applications. Springer; Singapore, Singapore: 2019. pp. 113–124. [Google Scholar]
- 90.Wu L., Gao X., Xia F., Joshi J., Borza T., Wang-Pruski G. Biostimulant and fungicidal effects of phosphite assessed by GC-TOF-MS analysis of potato leaf metabolome. Physiol. Mol. Plant Pathol. 2019;106:49–56. FAOSTAT (2019) http://www.fao.org/faostat/en/#data. Accessed 8 Mar 2019. [Google Scholar]