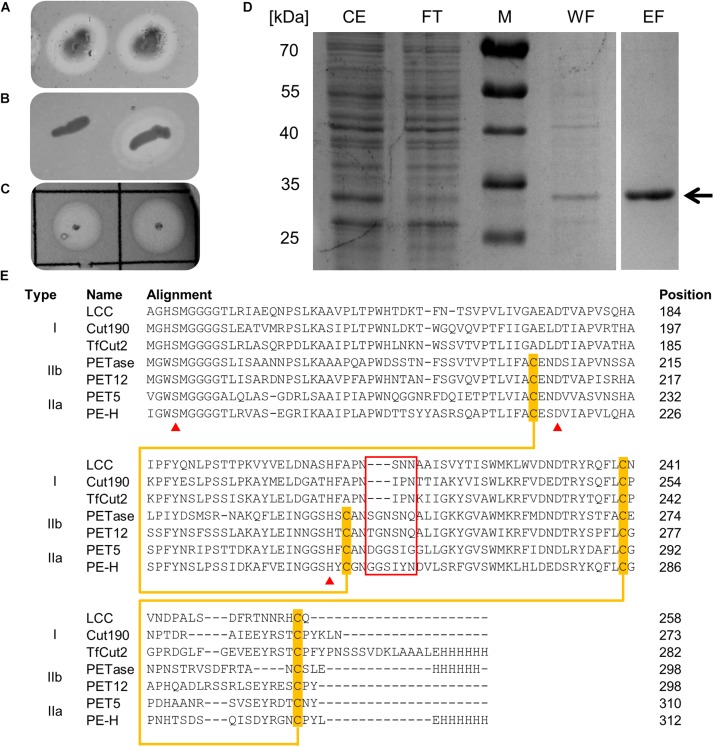
FIGURE 1.
(A) Colonies of P. aestusnigri grown on an Impranil DLN containing agar plate show clearing halos, indicating polyester hydrolase activity. (B) Hydrolytic activity of E. coli BL21(DE3) empty vector control (left) and PE-H production strain (right) on Impranil DLN containing agar plate. (C) Hydrolytic activity of purified PE-H on Impranil DLN containing agar plate. (D) Coomassie brilliant blue stained gel after SDS-PAGE of cell extracts of E. coli BL21(DE3) containing plasmid pET22b_PE-Hc6H before and after purification by IMAC; the position of PE-H is indicated by an arrow. Lanes contained cell extract (CE), flow through (FT), washing step (WF), and eluted protein (EF); the size of the molecular weight standards (M) are indicated on the left. (E) Part of a multiple sequence alignment of PE-H with amino acid sequences of different PET hydrolytic enzymes using the program Clustal Omega. The full length alignment can be found in the Supplementary Figure S1. The enzymes were assigned to different types of polyester hydrolases (Joo et al., 2018). Amino acid residues of the catalytic triad are marked by a red triangle, disulfide forming cysteine residues are highlighted in orange and connected by an orange line. Amino acids forming the extended loop region which is specific for type II PET hydrolytic enzymes are framed in red. Abbreviations are: leaf-branch compost metagenome cutinase (LCC); Saccharomonospora viridis cutinase (Cut190); Thermobifida fusca cutinase (TfCut2); Ideonella sakaiensis PET hydrolase (PETase); Polyangium brachysporum PET hydrolase (PET12); Oleispira antarctica PET hydrolase (PET5).