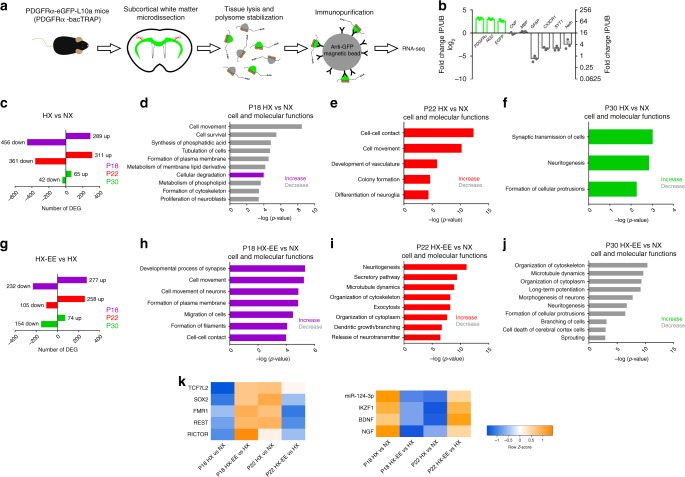
Fig. 8. RNA-seq reveals EE-induced changes in OPC translatome following perinatal hypoxia.
a Schematic representation of experimental design and TRAP methodology. b qRT-PCR analysis of immunoprecipitated (IP) and unbound (UB) fractions from the white matter of PDGFRɑ-bacTRAP mice. Green bars correspond to genes enriched in OPCs, and grayscale bars correspond to genes enriched in other cell types. Bars represent mean ± SEM (n = 3 per group). Platelet-derived growth factor receptor alpha (Pdgfrα), neural/glial antigen 2 (NG2), enhanced green fluorescent protein (Egfp), Cnp, myelin basic protein (Mbp), glial fibrillary acidic protein (Gfap; astrocytes), fractalkine receptor (Cx3cr1; microglia), synaptotagmin-1 (Syt1; neurons), and neurofilament-heavy (Nefh; axons). c Number of DEGs between HX and NX mice at P18, P22, and P30 (Wald test with Benjamini−Hochberg post hoc, adjusted p < 0.05, normalized counts > 5) (P18: HX, n = 4, NX, n = 3; P22: HX, n = 5, NX, n = 6; P30: HX, n = 3, NX, n = 6). d–f Predicted increases (colored) and decreases (gray) in cell and molecular functions (determined by directional z-scores) in HX compared to NX at P18 (d), P22 (e), and P30 (f). Cell and molecular functions ranked based on p value as determined using Fisher’s Exact Test. g Number of DEGs between HX-EE and HX mice at P18, P22, and P30 (Wald test with Benjamini−Hochberg post hoc, adjusted p < 0.05, normalized counts > 5) (P18: n = 4 per group; P22: n = 5 per group; P30: HX-EE, n = 5, HX, n = 3). h–j Predicted increases (colored) and decreases (gray) in cell and molecular functions (determined by directional z-scores) in HX-EE compared to HX at P18 (h), P22 (i), and P30 (j). Cell and molecular functions ranked based on p value as determined using Fisher’s Exact Test. k Heatmaps of most significantly predicted upstream regulators across four comparisons (P18 HX vs. NX, P18 HX-EE vs. HX, P22 HX vs. NX, and P22 HX-EE vs. HX). Boxes are colorized with z-scores (orange = activated, blue = inactivated). Source data are provided as a Source Data file.