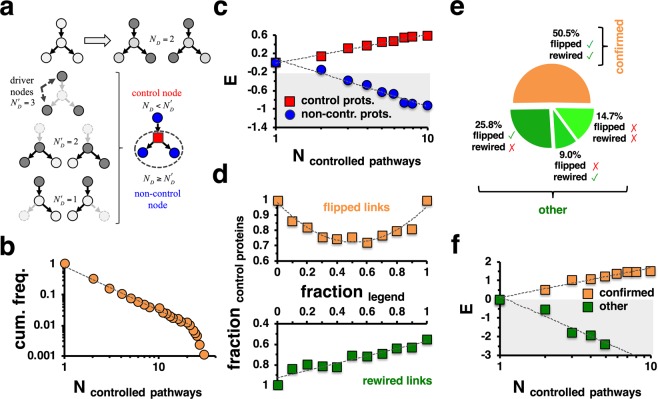
Figure 1.
Topological characteristics of pathway controlling proteins. (a) In the schematic representation of the controllability framework the application of a maximum matching algorithm allows the determination of driver nodes. To find control nodes, we separately eliminated each node and determined the number of driver nodes in the network thus obtained. We found a control node if the elimination of a node increased the number of driver nodes compared to the unperturbed network, . (b) Utilizing 276 KEGG pathways, we found that the cumulative frequency distribution of the number of pathways that proteins controlled followed a power-law, suggesting that a minority of proteins controls a large number of pathways and vice versa. (c) After determining control nodes in a network of 67,038 directed interactions between 5,398 proteins that we obtained by combining interactions of all pathways we randomly sampled such sets of control proteins. Notably, control proteins in the combined interaction network were enriched in groups of proteins that controlled an increasing number of pathways. (d) In the combined network we flipped and rewired given fractions of interactions. Notably, flipping the direction of roughly half of all interactions limited our ability to confirm control proteins the most. In turn, rewiring interactions continuously decreased the fraction of confirmed nodes. (e) When we flipped 50% of all interactions and rewired all interactions, respectively, half of all control proteins were confirmed. (f) More quantitatively, we randomly sampled sets of control proteins that were confirmed after flipping and rewiring interactions and found that such proteins were enriched in groups of proteins that controlled an increasing number of pathways. In turn, the set of remaining proteins was found strongly diluted in groups of proteins that controlled a limited number of pathways.