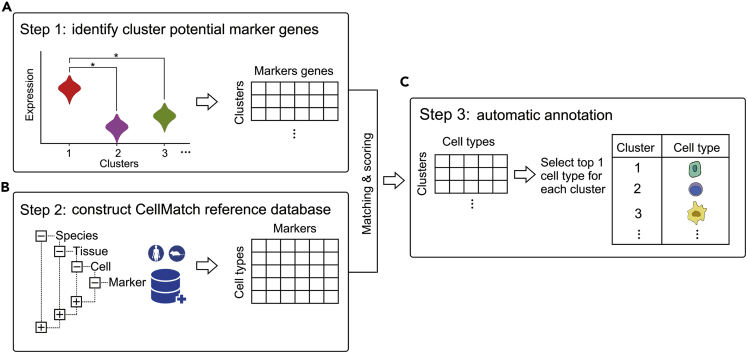
Figure 1.
Automatic Annotation on Cell Types of Clusters from scRNA-Seq Data Using scCATCH
(A) Paired comparison of clusters to identify the potential marker genes for each cluster. Compared with every other cluster, genes significantly upregulated in only one cluster (log10 fold change ≥0.25, p < 0.05) and expressed in more than a quarter of cells (≥25%) would be considered marker genes. p values were obtained through the Wilcoxon test. ∗ indicates p < 0.05.
(B) Construction of tissue-specific cell taxonomy reference databases (CellMatch) with tissue-specific cell markers reported in the literature from humans or mice.
(C) Evidence-based score and annotation. For each cluster, cell types were scored on the basis of validated marker genes and their supporting literature, and the cell type with the highest score (top 1) was determined for the cluster.