Figure 3.
Comparison of scCATCH with Other Methods
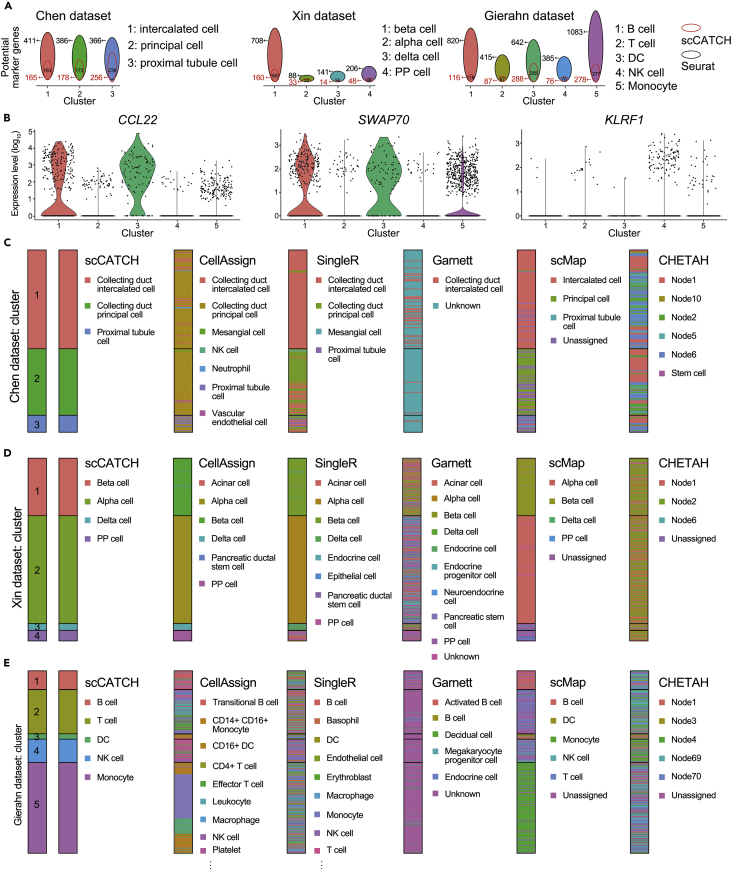
(A) Identification of cluster potential marker genes via Seurat (black number beside the circle) and scCATCH (red numbers beside the circle) in three validation datasets in each cluster. The black number inside the circle represents the number of overlapped genes.
(B) The violin plot for the expression levels of cluster 1 marker gene CCL22, cluster 3 marker gene SWAP70, and cluster 4 marker gene KLRF1 across 5 clusters identified in the Gierahn dataset via Seurat.
(C) Cell type annotation in the Chen dataset via scCATCH and cell-based annotation including CellAssign, SingleR, Garnett, scMap, and CHETAH.
(D) Cell type annotation in the Xin dataset via scCATCH, CellAssign, SingleR, Garnett, scMap, and CHETAH.
(E) Cell type annotation of the Gierahn dataset. The representative cell types were labeled for CellAssign and SingleR. Nodes 1, 2, 3, 4, 5, 6, 10, 69, and 70 represent intermediate cell types with a low confidence score.