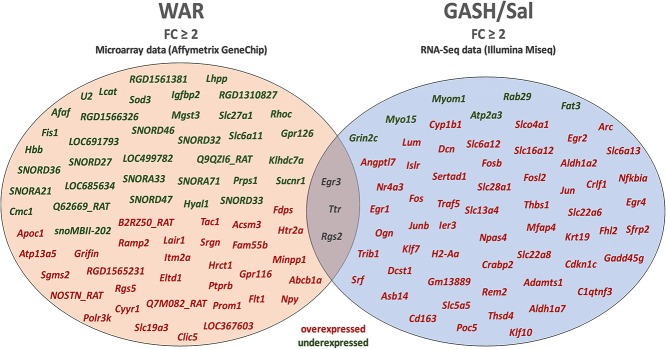
Figure 1.
Diagram with the genes identified as differentially regulated in the inferior colliculus (IC) of the WAR and GASH/Sal models using the selection criterion of fold change (|FC| ≥ 2). cDNA microarray analysis and RNA-Seq identified a total of 71 and 64 differentially expressed genes in the WAR and GASH/Sal, respectively. A set of three differentially expressed genes (Egr3, Ttr, and Rgs2) overlapped between the microarray and RNA-Seq approaches. Genes highlighted in red: overexpressed genes; Genes highlighted in green: underexpressed genes; Genes highlighted in gray: common differentially expressed genes.