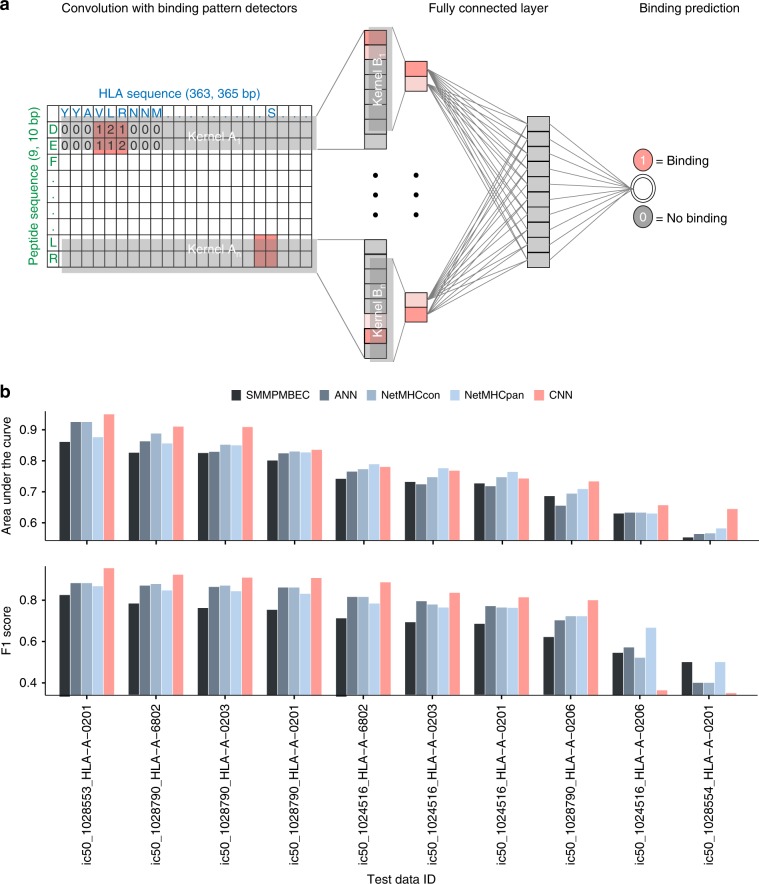
Fig. 1. Prediction model for peptide-MHC class I binding and performance evaluation.
a CNN architecture was used to predict binding between peptides and MHC class I molecules. The two-dimensional map of interactions between amino acids in peptide-MHC class I complex was used as the input matrix. A set of kernels, A1,…An, covering the entire HLA sequence were applied on the input matrix. The output convolution scores in the first layer were scanned by the second set of kernels, B1,…, Bn. A fully connected layer attached to the second layer integrated the convoluted patterns for classification. b Comparison of prediction performance with SMMPMBEC, artificial neural network (ANN), NetMHCcon, and NetMHCpan on the basis of weekly updated test datasets of IEDB. In terms of AUC, our method was superior to SMMPMBEC, ANN, NetMHCcon, and NetMHCpan for 100%, 100%, 90%, and 70% of the test cases. With regards to the F1 score, our CNN was superior to all the methods for 80% of the test cases. Source data are provided as a Source Data file.