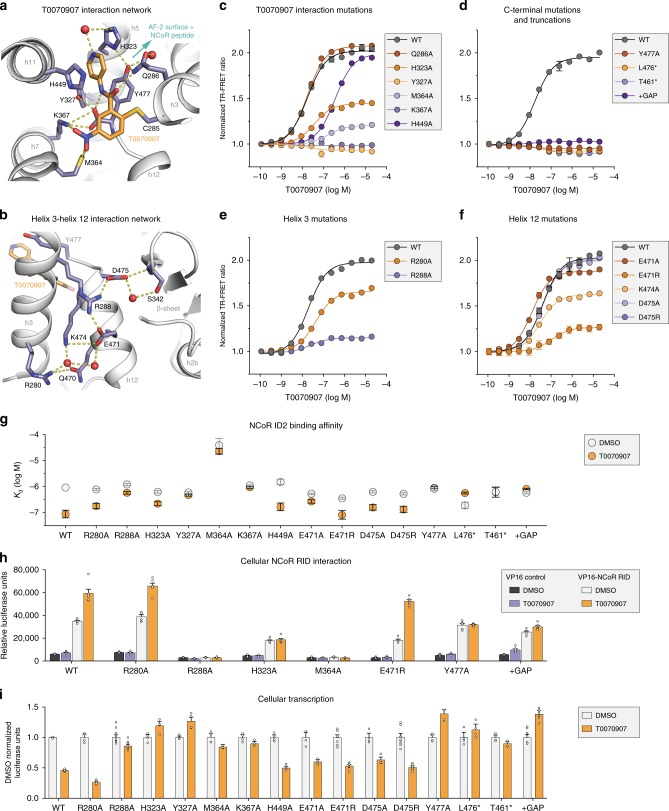
Fig. 6. Mutagenesis validates the inverse agonist-dependent repressive helix 12 conformation.
a Interactions between PPARγ and the corepressor-selective inverse agonist T0070907 (PDB 6ONI). b The repressive helix 12 conformation is mediated through a network of interactions with residues on helix 3 (PDB 6ONI). c, d Structure-guided mutants of the T0070907 interaction tested using a TR-FRET biochemical assay measuring the ligand-dependent change in the interaction of PPARγ LBD with a peptide derived from the NCoR corepressor (n = 3; mean ± s.e.m.). e, f Structure-guided mutants of the helix 3/helix 12 interaction network tested using the TR-FRET biochemical assay (n = 3; mean ± s.e.m.). g NCoR ID2 peptide affinities and errors (mean ± s.d. of the fit to a one site—total binding equation) from fluorescence polarization peptide interaction assays using structure-guided mutants (Supplementary Fig. 4) displayed to illustrate the difference in NCoR ID2 peptide binding affinity between apo- and T0070907-bound PPARγ LBD. h Mammalian two-hybrid assay measuring the effect of T0070907 (10 μM) on the interaction between the Gal4-PPARγ LBD and the VP16-NCoR RID (n = 6; mean ± s.e.m.). i Luciferase transcriptional reporter assay in HEK293T cells treated with DMSO control or 5 μM T0070907 measuring the ligand-dependent change in wild-type or mutant PPARγ transcription; data normalized to each WT or mutant construct vehicle control (DMSO) condition (n = 4–12; mean ± s.e.m.). Source data are provided as a Source Data file.